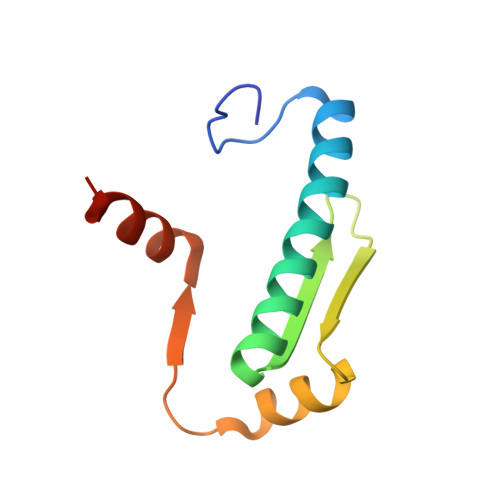
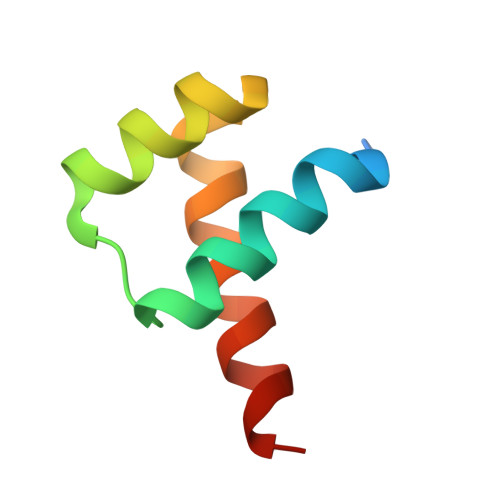
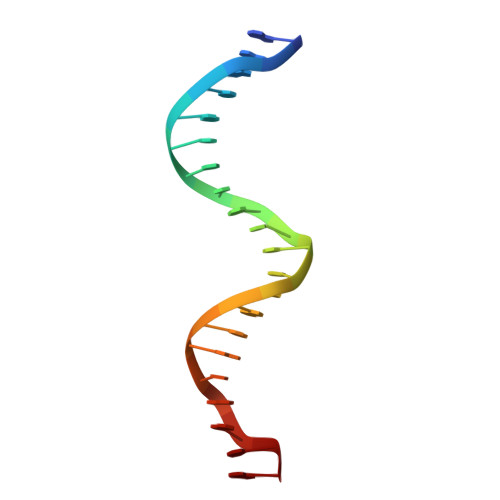
Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
Lei, X., Zhao, J., Sagendorf, J.M., Rajashekar, N., Xu, J., Dantas Machado, A.C., Sen, C., Rohs, R., Feng, P., Chen, L.(2020) J Mol Biology 432: 5499-5508
- PubMed: 32681840
- DOI: https://doi.org/10.1016/j.jmb.2020.07.004
- Primary Citation of Related Structures:
6WC2, 6WC5 - PubMed Abstract:
MEF2 and NKX2-5 transcription factors interact with each other in cardiogenesis and are necessary for normal heart formation. Despite evidence suggesting that these two transcription factors function synergistically and possibly through direct physical interactions, molecular mechanisms by which they interact are not clear. Here we determined the crystal structures of ternary complexes of MEF2 and NKX2-5 bound to myocardin enhancer DNA in two crystal forms. These crystal structures are the first example of human MADS-box/homeobox ternary complex structures involved in cardiogenesis. Our structures reveal two possible modes of interactions between MEF2 and NKX2-5: MEF2 and NKX bind to adjacent DNA sites to recognize DNA in cis; and MEF2 and NKX bind to different DNA strands to interact with each other in trans via a conserved protein-protein interface observed in both crystal forms. Disease-related mutations are mapped to the observed protein-protein interface. Our structural studies provide a starting point to understand and further study the molecular mechanisms of the interactions between MEF2 and NKX2.5 and their roles in cardiogenesis.
- Molecular and Computational Biology, Department of Biological Sciences, University of Southern California, Los Angeles, CA 90089, USA. Electronic address: xiaoleiusc@gmail.com.
Organizational Affiliation: