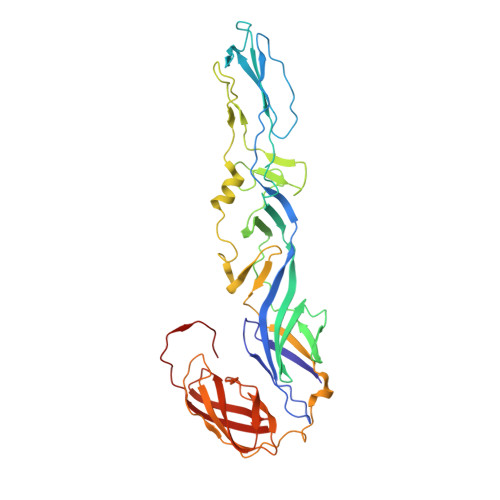
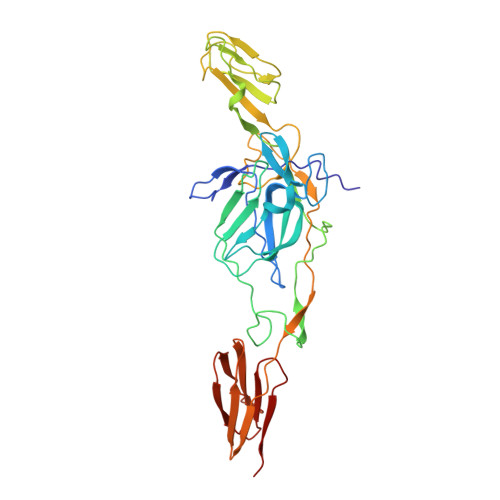
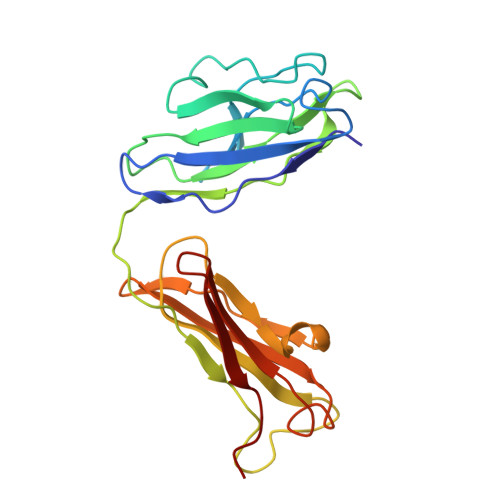
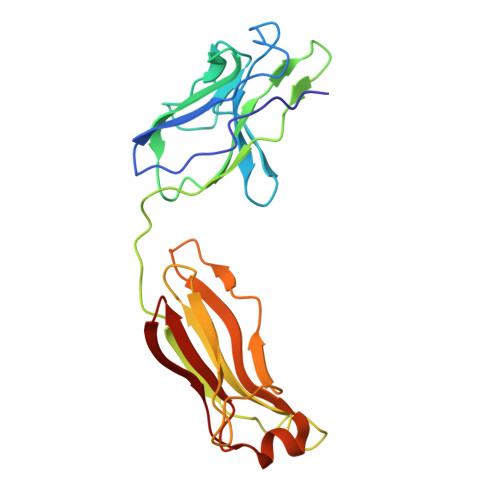
Human mAbs Broadly Protect against Arthritogenic Alphaviruses by Recognizing Conserved Elements of the Mxra8 Receptor-Binding Site.
Powell, L.A., Miller, A., Fox, J.M., Kose, N., Klose, T., Kim, A.S., Bombardi, R., Tennekoon, R.N., Dharshan de Silva, A., Carnahan, R.H., Diamond, M.S., Rossmann, M.G., Kuhn, R.J., Crowe Jr., J.E.(2020) Cell Host Microbe 28: 699-711.e7
- PubMed: 32783883
- DOI: https://doi.org/10.1016/j.chom.2020.07.008
- Primary Citation of Related Structures:
6VYV, 6W09, 6W1C, 6W2U - PubMed Abstract:
Mosquito inoculation of humans with arthritogenic alphaviruses results in a febrile syndrome characterized by debilitating musculoskeletal pain and arthritis. Despite an expanding global disease burden, no approved therapies or licensed vaccines exist. Here, we describe human monoclonal antibodies (mAbs) that bind to and neutralize multiple distantly related alphaviruses. These mAbs compete for an antigenic site and prevent attachment to the recently discovered Mxra8 alphavirus receptor. Three cryoelectron microscopy structures of Fab in complex with Ross River (RRV), Mayaro, or chikungunya viruses reveal a conserved footprint of the broadly neutralizing mAb RRV-12 in a region of the E2 glycoprotein B domain. This mAb neutralizes virus in vitro by preventing virus entry and spread and is protective in vivo in mouse models. Thus, the RRV-12 mAb and its defined epitope have potential as a therapeutic agent or target of vaccine design against multiple emerging arthritogenic alphavirus infections.
- Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, TN 37232, USA.
Organizational Affiliation: