Crystal structure of prephenate dehydratase from Brucella melitensis biovar abortus 2308 in complex with phenylalanine
Abendroth, J., Dranow, D.M., Lorimer, D.D., Bullen, J.C., Sankaran, B., Horanyi, P.S., Edwards, T.E.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Prephenate dehydratase:Amino acid-binding ACT | 308 | Brucella abortus 2308 | Mutation(s): 0 Gene Names: pheA, BAB1_0034 EC: 4.2.1.51 | 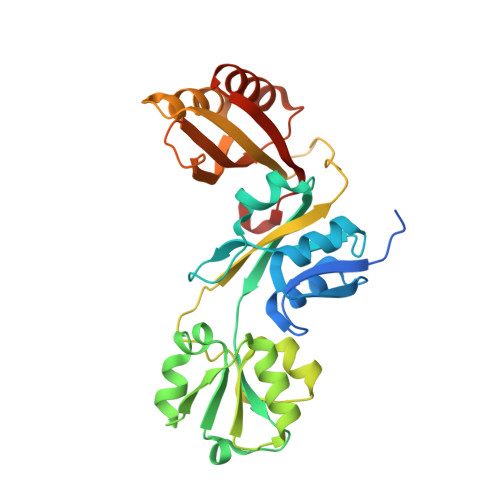 | |
UniProt | |||||
Find proteins for Q2YPM5 (Brucella abortus (strain 2308)) Explore Q2YPM5 Go to UniProtKB: Q2YPM5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2YPM5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PHE Query on PHE | E [auth A], G [auth B], J [auth C], L [auth D] | PHENYLALANINE C9 H11 N O2 COLNVLDHVKWLRT-QMMMGPOBSA-N |  | ||
| EDO Query on EDO | F [auth A] H [auth B] I [auth B] K [auth C] M [auth D] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 226.95 | α = 90 |
| b = 61.25 | β = 112.95 |
| c = 120.05 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| MR-Rosetta | phasing |