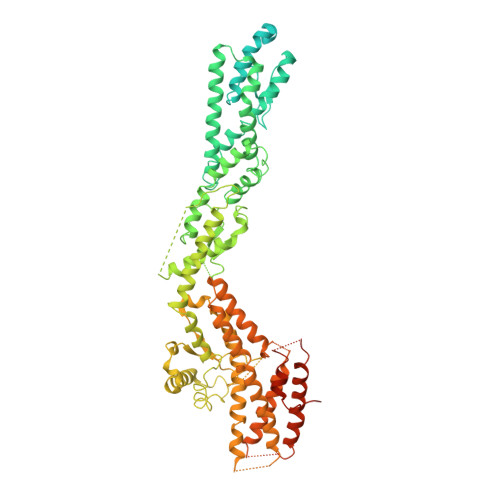
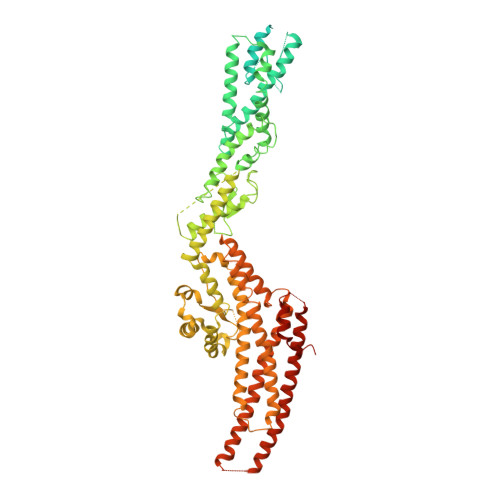
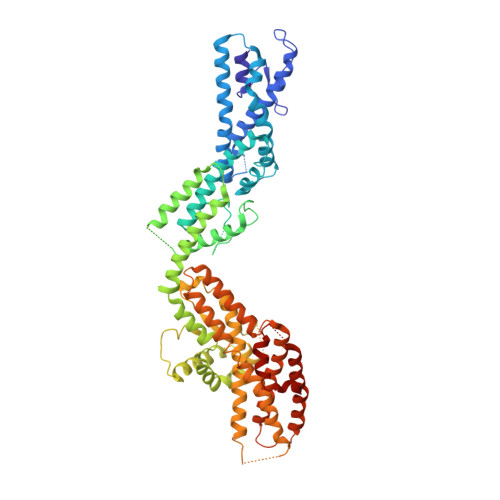
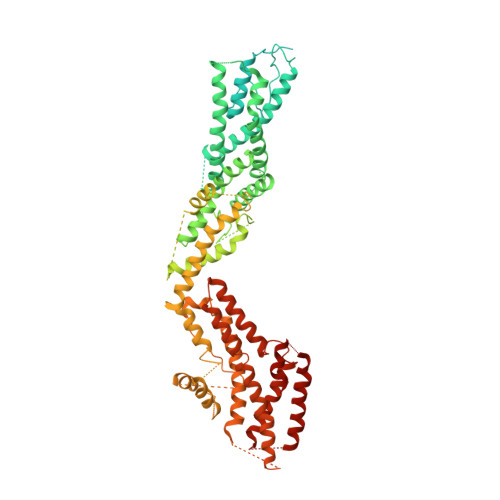
Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Wieczorek, M., Urnavicius, L., Ti, S.C., Molloy, K.R., Chait, B.T., Kapoor, T.M.(2020) Cell 180: 165-175.e16
- PubMed: 31862189
- DOI: https://doi.org/10.1016/j.cell.2019.12.007
- Primary Citation of Related Structures:
6V5V, 6V69, 6V6B, 6V6C, 6V6S - PubMed Abstract:
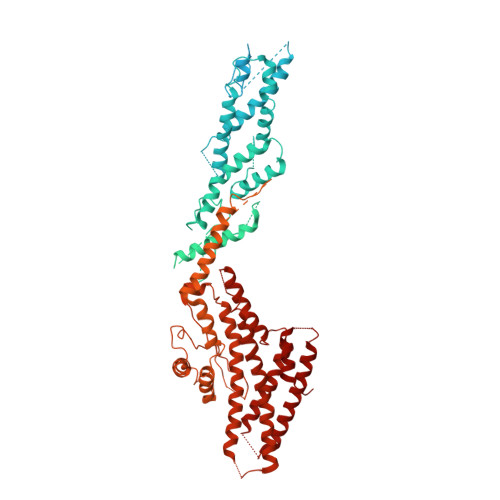
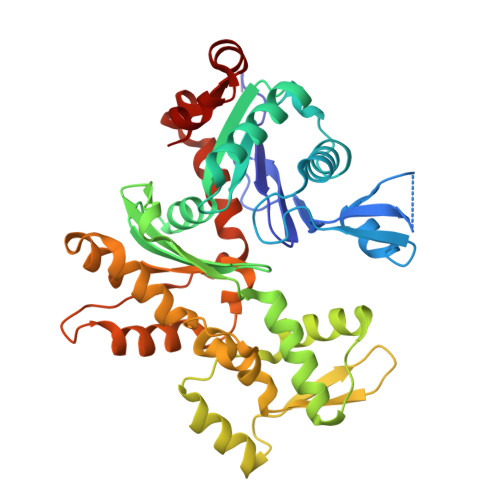
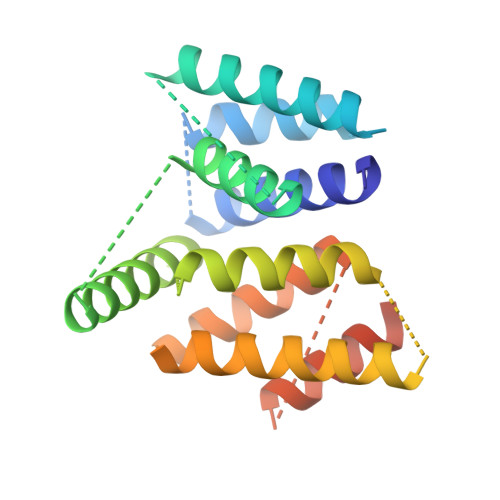
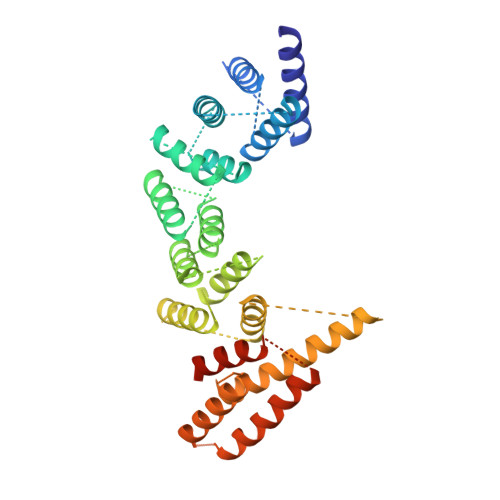
The γ-tubulin ring complex (γ-TuRC) is an essential regulator of centrosomal and acentrosomal microtubule formation, yet its structure is not known. Here, we present a cryo-EM reconstruction of the native human γ-TuRC at ∼3.8 Å resolution, revealing an asymmetric, cone-shaped structure. Pseudo-atomic models indicate that GCP4, GCP5, and GCP6 form distinct Y-shaped assemblies that structurally mimic GCP2/GCP3 subcomplexes distal to the γ-TuRC "seam." We also identify an unanticipated structural bridge that includes an actin-like protein and spans the γ-TuRC lumen. Despite its asymmetric architecture, the γ-TuRC arranges γ-tubulins into a helical geometry poised to nucleate microtubules. Diversity in the γ-TuRC subunits introduces large (>100,000 Å 2 ) surfaces in the complex that allow for interactions with different regulatory factors. The observed compositional complexity of the γ-TuRC could self-regulate its assembly into a cone-shaped structure to control microtubule formation across diverse contexts, e.g., within biological condensates or alongside existing filaments.
- Laboratory of Chemistry and Cell Biology, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.
Organizational Affiliation: