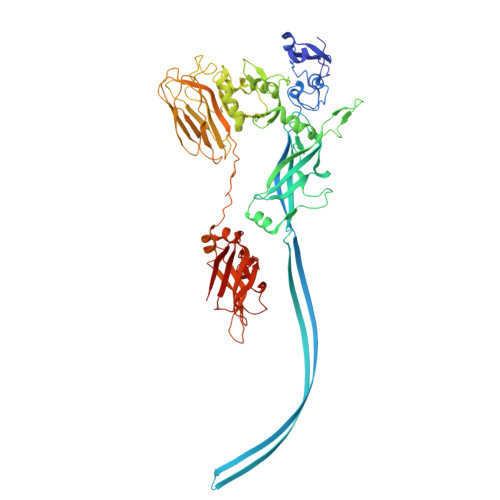
Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Xu, X., Godoy-Ruiz, R., Adipietro, K.A., Peralta, C., Ben-Hail, D., Varney, K.M., Cook, M.E., Roth, B.M., Wilder, P.T., Cleveland, T., Grishaev, A., Neu, H.M., Michel, S.L.J., Yu, W., Beckett, D., Rustandi, R.R., Lancaster, C., Loughney, J.W., Kristopeit, A., Christanti, S., Olson, J.W., MacKerell, A.D., Georges, A.D., Pozharski, E., Weber, D.J.(2020) Proc Natl Acad Sci U S A 117: 1049-1058
- PubMed: 31896582
- DOI: https://doi.org/10.1073/pnas.1919490117
- Primary Citation of Related Structures:
6UWI, 6UWO, 6UWR, 6UWT - PubMed Abstract:
Targeting Clostridium difficile infection is challenging because treatment options are limited, and high recurrence rates are common. One reason for this is that hypervirulent C. difficile strains often have a binary toxin termed the C. difficile toxin, in addition to the enterotoxins TsdA and TsdB. The C. difficile toxin has an enzymatic component, termed CDTa, and a pore-forming or delivery subunit termed CDTb. CDTb was characterized here using a combination of single-particle cryoelectron microscopy, X-ray crystallography, NMR, and other biophysical methods. In the absence of CDTa, 2 di-heptamer structures for activated CDTb (1.0 MDa) were solved at atomic resolution, including a symmetric ( Sym CDTb; 3.14 Å) and an asymmetric form ( Asym CDTb; 2.84 Å). Roles played by 2 receptor-binding domains of activated CDTb were of particular interest since the receptor-binding domain 1 lacks sequence homology to any other known toxin, and the receptor-binding domain 2 is completely absent in other well-studied heptameric toxins (i.e., anthrax). For Asym CDTb, a Ca 2+ binding site was discovered in the first receptor-binding domain that is important for its stability, and the second receptor-binding domain was found to be critical for host cell toxicity and the di-heptamer fold for both forms of activated CDTb. Together, these studies represent a starting point for developing structure-based drug-design strategies to target the most severe strains of C. difficile .
- City University of New York Advanced Science Research Center, City University of New York, New York, NY 10017.
Organizational Affiliation: