Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Wang, R., Qi, X., Schmiege, P., Coutavas, E., Li, X.(2020) Sci Adv 6: eaaz1466
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2020) Sci Adv 6: eaaz1466
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cation-independent mannose-6-phosphate receptor | 2,499 | Bos taurus | Mutation(s): 0 | 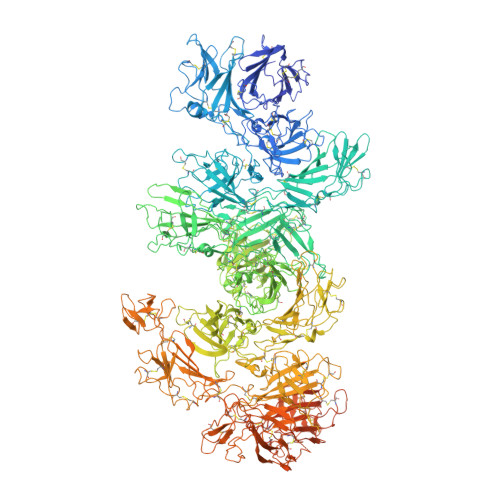 | |
UniProt | |||||
Find proteins for P08169 (Bos taurus) Explore P08169 Go to UniProtKB: P08169 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08169 | ||||
Glycosylation | |||||
| Glycosylation Sites: 4 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | D [auth A], E [auth A], F [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||