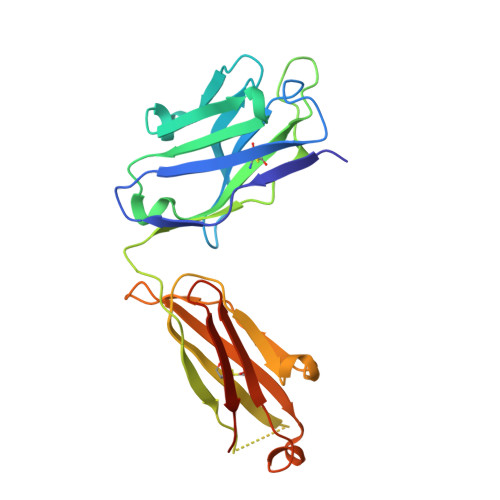
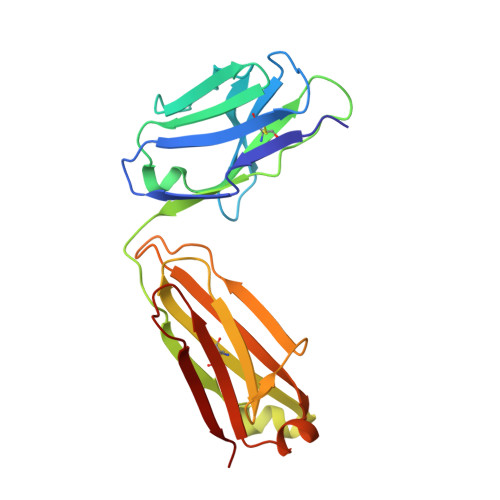
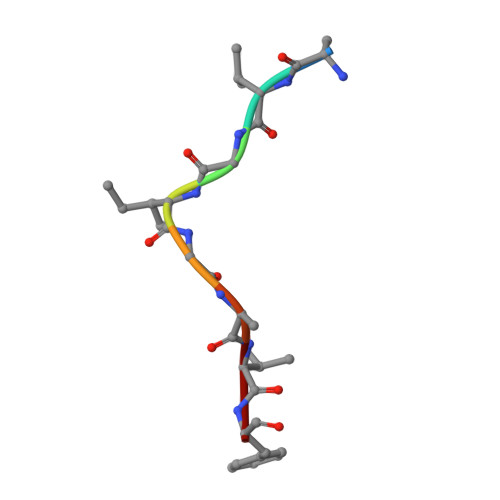
VRC34-Antibody Lineage Development Reveals How a Required Rare Mutation Shapes the Maturation of a Broad HIV-Neutralizing Lineage.
Shen, C.H., DeKosky, B.J., Guo, Y., Xu, K., Gu, Y., Kilam, D., Ko, S.H., Kong, R., Liu, K., Louder, M.K., Ou, L., Zhang, B., Chao, C.W., Corcoran, M.M., Feng, E., Huang, J., Normandin, E., O'Dell, S., Ransier, A., Rawi, R., Sastry, M., Schmidt, S.D., Wang, S., Wang, Y., Chuang, G.Y., Doria-Rose, N.A., Lin, B., Zhou, T., Boritz, E.A., Connors, M., Douek, D.C., Karlsson Hedestam, G.B., Sheng, Z., Shapiro, L., Mascola, J.R., Kwong, P.D.(2020) Cell Host Microbe 27: 531
- PubMed: 32130953
- DOI: https://doi.org/10.1016/j.chom.2020.01.027
- Primary Citation of Related Structures:
6UBI, 6UCE, 6UCF - PubMed Abstract:
Rare mutations have been proposed to restrict the development of broadly neutralizing antibodies against HIV-1, but this has not been explicitly demonstrated. We hypothesized that such rare mutations might be identified by comparing broadly neutralizing and non-broadly neutralizing branches of an antibody-developmental tree. Because sequences of antibodies isolated from the fusion peptide (FP)-targeting VRC34-antibody lineage suggested it might be suitable for such rare mutation analysis, we carried out next-generation sequencing (NGS) on B cell transcripts from donor N123, the source of the VRC34 lineage, and functionally and structurally characterized inferred intermediates along broadly neutralizing and poorly neutralizing developmental branches. The broadly neutralizing VRC34.01 branch required the rare heavy-chain mutation Y33P to bind FP, whereas the early bifurcated VRC34.05 branch did not require this rare mutation and evolved less breadth. Our results demonstrate how a required rare mutation can restrict development and shape the maturation of a broad HIV-1-neutralizing antibody lineage.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: