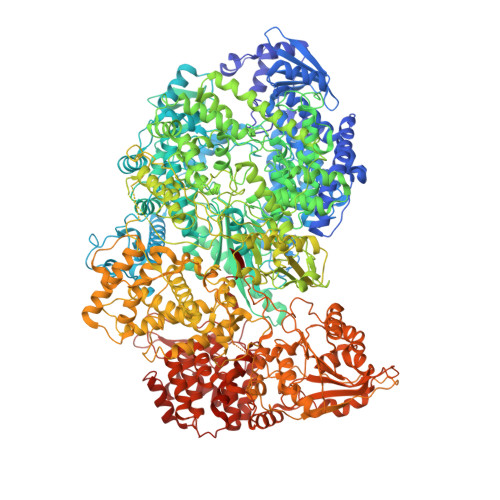
Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Jenni, S., Bloyet, L.M., Diaz-Avalos, R., Liang, B., Whelan, S.P.J., Grigorieff, N., Harrison, S.C.(2020) Cell Rep 30: 53-60.e5
- PubMed: 31914397
- DOI: https://doi.org/10.1016/j.celrep.2019.12.024
- Primary Citation of Related Structures:
6U1X - PubMed Abstract:
The large (L) proteins of non-segmented, negative-strand RNA viruses are multifunctional enzymes that produce capped, methylated, and polyadenylated mRNA and replicate the viral genome. A phosphoprotein (P), required for efficient RNA-dependent RNA polymerization from the viral ribonucleoprotein (RNP) template, regulates the function and conformation of the L protein. We report the structure of vesicular stomatitis virus L in complex with its P cofactor determined by electron cryomicroscopy at 3.0 Å resolution, enabling us to visualize bound segments of P. The contacts of three P segments with multiple L domains show how P induces a closed, compact, initiation-competent conformation. Binding of P to L positions its N-terminal domain adjacent to a putative RNA exit channel for efficient encapsidation of newly synthesized genomes with the nucleoprotein and orients its C-terminal domain to interact with an RNP template. The model shows that a conserved tryptophan in the priming loop can support the initiating 5' nucleotide.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: