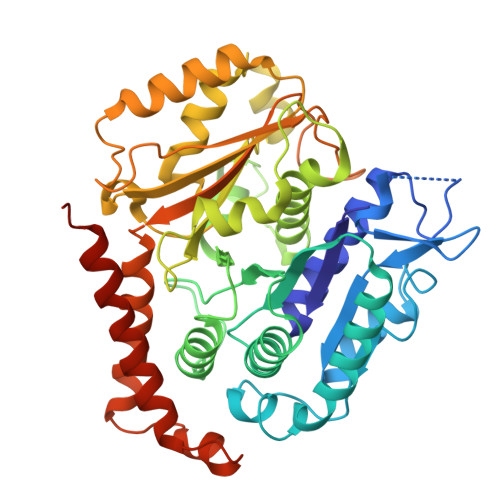
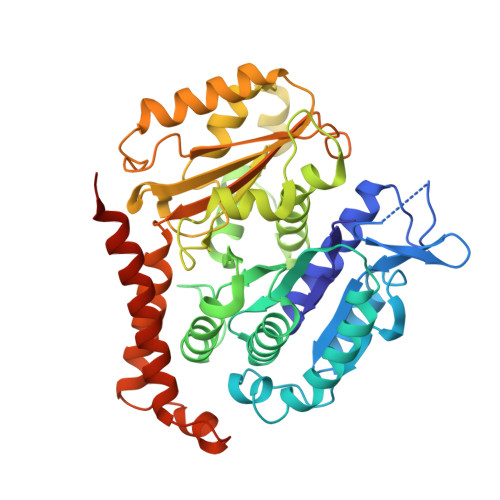
Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins.
Ichikawa, M., Khalifa, A.A.Z., Kubo, S., Dai, D., Basu, K., Maghrebi, M.A.F., Vargas, J., Bui, K.H.(2019) Proc Natl Acad Sci U S A 116: 19930-19938
- PubMed: 31527277
- DOI: https://doi.org/10.1073/pnas.1911119116
- Primary Citation of Related Structures:
6U0H, 6U0T, 6U0U - PubMed Abstract:
Cilia, the hair-like protrusions that beat at high frequencies to propel a cell or move fluid around are composed of radially bundled doublet microtubules. In this study, we present a near-atomic resolution map of the Tetrahymena doublet microtubule by cryoelectron microscopy. The map demonstrates that the network of microtubule inner proteins weaves into the tubulin lattice and forms an inner sheath. From mass spectrometry data and de novo modeling, we identified Rib43a proteins as the filamentous microtubule inner proteins in the protofilament ribbon region. The Rib43a-tubulin interaction leads to an elongated tubulin dimer distance every 2 dimers. In addition, the tubulin lattice structure with missing microtubule inner proteins (MIPs) by sarkosyl treatment shows significant longitudinal compaction and lateral angle change between protofilaments. These results are evidence that the MIPs directly affect and stabilize the tubulin lattice. It suggests that the doublet microtubule is an intrinsically stressed filament and that this stress could be manipulated in the regulation of ciliary waveforms.
- Department of Anatomy and Cell Biology, McGill University, Montréal, QC H3A 0C7, Canada.
Organizational Affiliation: