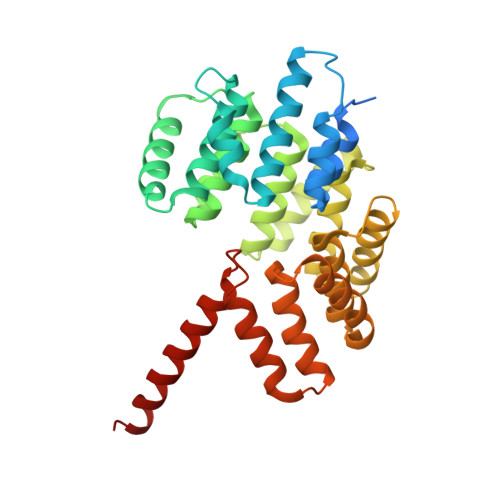
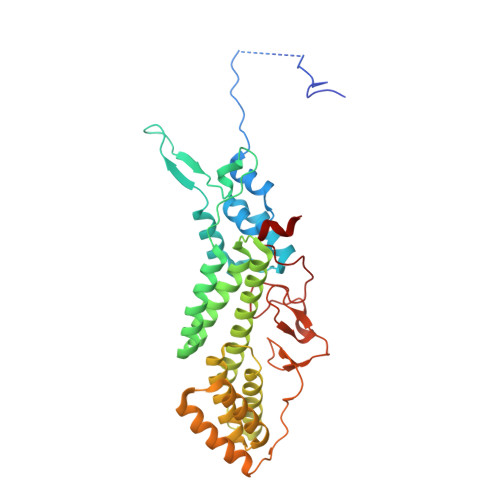
Roles of singleton tryptophan motifs in COPI coat stability and vesicle tethering.
Travis, S.M., Kokona, B., Fairman, R., Hughson, F.M.(2019) Proc Natl Acad Sci U S A 116: 24031-24040
- PubMed: 31712447
- DOI: https://doi.org/10.1073/pnas.1909697116
- Primary Citation of Related Structures:
6TZT, 6U3V, 6U3W - PubMed Abstract:
Coat protein I (COPI)-coated vesicles mediate retrograde transport from the Golgi to the endoplasmic reticulum (ER), as well as transport within the Golgi. Major progress has been made in defining the structure of COPI coats, in vitro and in vivo, at resolutions as high as 9 Å. Nevertheless, important questions remain unanswered, including what specific interactions stabilize COPI coats, how COPI vesicles recognize their target membranes, and how coat disassembly is coordinated with vesicle fusion and cargo delivery. Here, we use X-ray crystallography to identify a conserved site on the COPI subunit α-COP that binds to flexible, acidic sequences containing a single tryptophan residue. One such sequence, found within α-COP itself, mediates α-COP homo-oligomerization. Another such sequence is contained within the lasso of the ER-resident Dsl1 complex, where it helps mediate the tethering of Golgi-derived COPI vesicles at the ER membrane. Together, our findings suggest that α-COP homo-oligomerization plays a key role in COPI coat stability, with potential implications for the coordination of vesicle tethering, uncoating, and fusion.
- Department of Molecular Biology, Princeton University, Princeton, NJ 08544.
Organizational Affiliation: