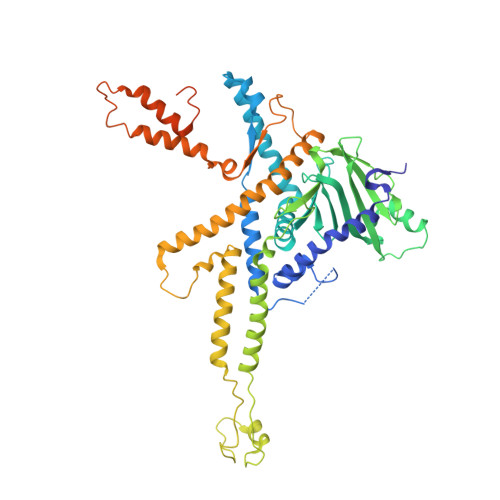
Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Fabrega-Ferrer, M., Cuervo, A., Fernandez, F.J., Machon, C., Perez-Luque, R., Pous, J., Vega, M.C., Carrascosa, J.L., Coll, M.(2021) Acta Crystallogr D Struct Biol 77: 11-18
- PubMed: 33404521
- DOI: https://doi.org/10.1107/S2059798320015156
- Primary Citation of Related Structures:
6TJP - PubMed Abstract:
Medium-resolution cryo-electron microscopy maps, in particular when they include a significant number of α-helices, may allow the building of partial models that are useful for molecular-replacement searches in large crystallographic structures when the structures of homologs are not available and experimental phasing has failed. Here, as an example, the solution of the structure of a bacteriophage portal using a partial 30% model built into a 7.8 Å resolution cryo-EM map is shown. Inspection of the self-rotation function allowed the correct oligomerization state to be determined, and density-modification procedures using rotation matrices and a mask based on the cryo-EM structure were critical for solving the structure. A workflow is described that may be applicable to similar cases and this strategy is compared with direct use of the cryo-EM map for molecular replacement.
- Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain.
Organizational Affiliation: