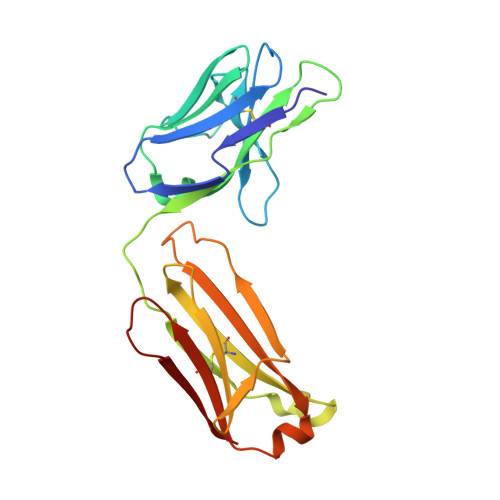
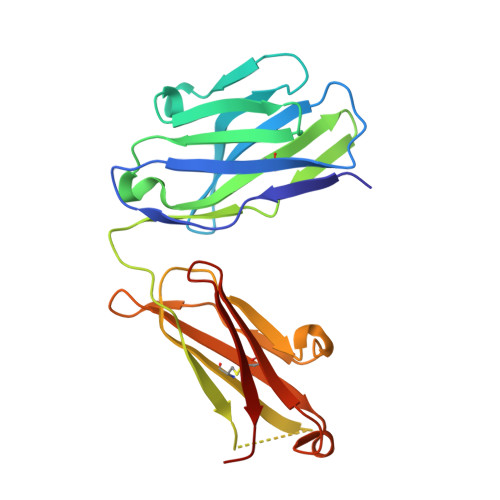
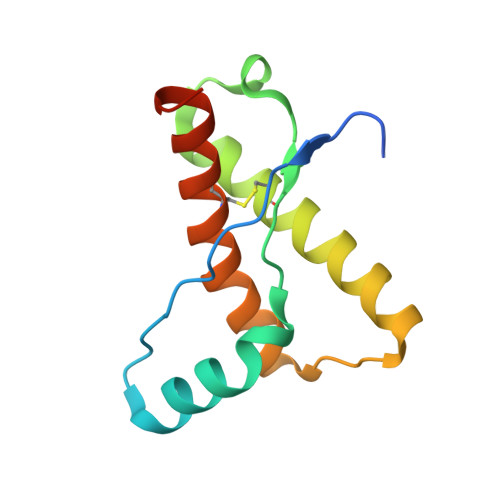
Structural effects of the highly protective V127 polymorphism on human prion protein.
Hosszu, L.L.P., Conners, R., Sangar, D., Batchelor, M., Sawyer, E.B., Fisher, S., Cliff, M.J., Hounslow, A.M., McAuley, K., Leo Brady, R., Jackson, G.S., Bieschke, J., Waltho, J.P., Collinge, J.(2020) Commun Biol 3: 402-402
- PubMed: 32728168
- DOI: https://doi.org/10.1038/s42003-020-01126-6
- Primary Citation of Related Structures:
6SUZ, 6SV2 - PubMed Abstract:
Prion diseases, a group of incurable, lethal neurodegenerative disorders of mammals including humans, are caused by prions, assemblies of misfolded host prion protein (PrP). A single point mutation (G127V) in human PrP prevents prion disease, however the structural basis for its protective effect remains unknown. Here we show that the mutation alters and constrains the PrP backbone conformation preceding the PrP β-sheet, stabilising PrP dimer interactions by increasing intermolecular hydrogen bonding. It also markedly changes the solution dynamics of the β2-α2 loop, a region of PrP structure implicated in prion transmission and cross-species susceptibility. Both of these structural changes may affect access to protein conformers susceptible to prion formation and explain its profound effect on prion disease.
- MRC Prion Unit at UCL, UCL Institute of Prion Diseases, 33 Cleveland Street, London, W1W 7FF, UK.
Organizational Affiliation: