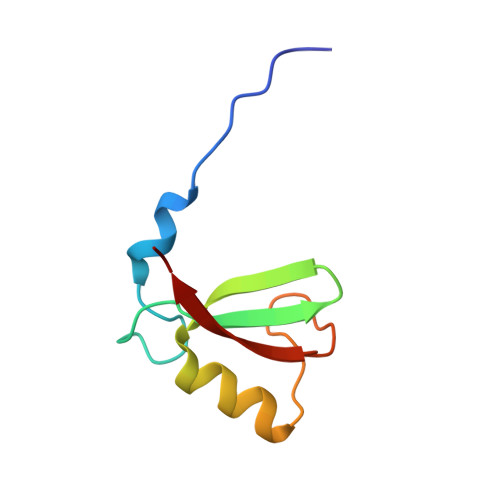
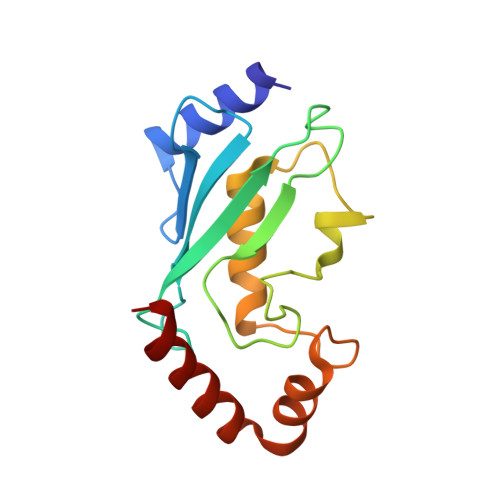
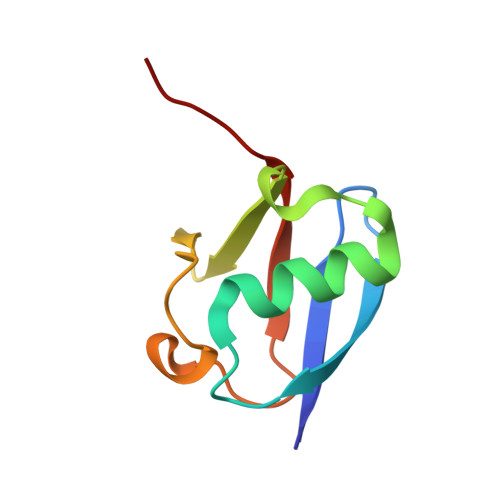
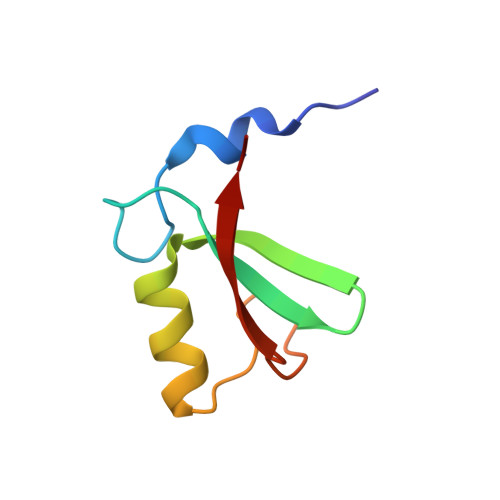
Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Magnussen, H.M., Ahmed, S.F., Sibbet, G.J., Hristova, V.A., Nomura, K., Hock, A.K., Archibald, L.J., Jamieson, A.G., Fushman, D., Vousden, K.H., Weissman, A.M., Huang, D.T.(2020) Nat Commun 11: 2094-2094
- PubMed: 32350255
- DOI: https://doi.org/10.1038/s41467-020-15783-y
- Primary Citation of Related Structures:
6SQO, 6SQP, 6SQR, 6SQS - PubMed Abstract:
Phosphorylation of MDM2 by ATM upon DNA damage is an important mechanism for deregulating MDM2, thereby leading to p53 activation. ATM phosphorylates multiple residues near the RING domain of MDM2, but the underlying molecular basis for deregulation remains elusive. Here we show that Ser429 phosphorylation selectively enhances the ubiquitin ligase activity of MDM2 homodimer but not MDM2-MDMX heterodimer. A crystal structure of phospho-Ser429 (pS429)-MDM2 bound to E2-ubiquitin reveals a unique 3 10 -helical feature present in MDM2 homodimer that allows pS429 to stabilize the closed E2-ubiquitin conformation and thereby enhancing ubiquitin transfer. In cells Ser429 phosphorylation increases MDM2 autoubiquitination and degradation upon DNA damage, whereas S429A substitution protects MDM2 from auto-degradation. Our results demonstrate that Ser429 phosphorylation serves as a switch to boost the activity of MDM2 homodimer and promote its self-destruction to enable rapid p53 stabilization and resolve a long-standing controversy surrounding MDM2 auto-degradation in response to DNA damage.
- Cancer Research UK Beatson Institute, Garscube Estate, Switchback Road, Glasgow, G61 1BD, UK.
Organizational Affiliation: