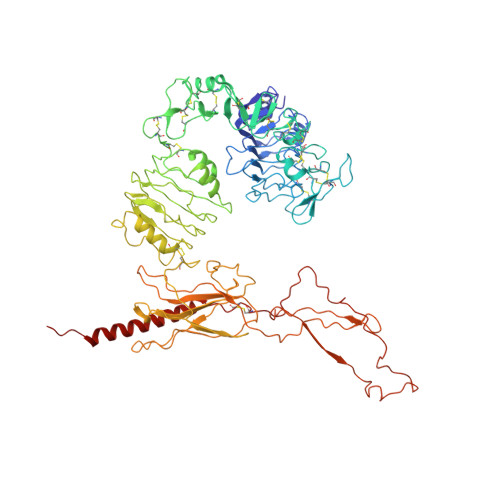
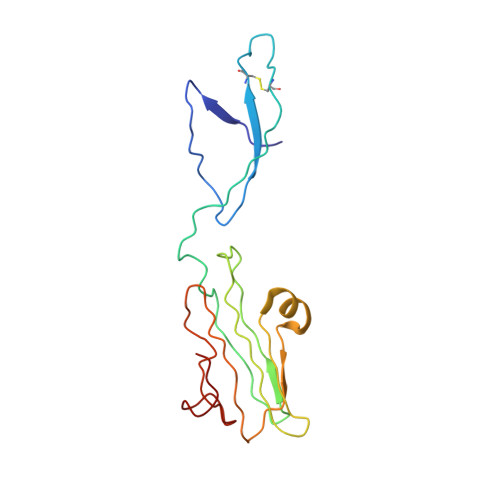
Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
Gutmann, T., Schafer, I.B., Poojari, C., Brankatschk, B., Vattulainen, I., Strauss, M., Coskun, U.(2020) J Cell Biol 219
- PubMed: 31727777
- DOI: https://doi.org/10.1083/jcb.201907210
- Primary Citation of Related Structures:
6SOF, 7QID - PubMed Abstract:
Glucose homeostasis and growth essentially depend on the hormone insulin engaging its receptor. Despite biochemical and structural advances, a fundamental contradiction has persisted in the current understanding of insulin ligand-receptor interactions. While biochemistry predicts two distinct insulin binding sites, 1 and 2, recent structural analyses have resolved only site 1. Using a combined approach of cryo-EM and atomistic molecular dynamics simulation, we present the structure of the entire dimeric insulin receptor ectodomain saturated with four insulin molecules. Complementing the previously described insulin-site 1 interaction, we present the first view of insulin bound to the discrete insulin receptor site 2. Insulin binding stabilizes the receptor ectodomain in a T-shaped conformation wherein the membrane-proximal domains converge and contact each other. These findings expand the current models of insulin binding to its receptor and of its regulation. In summary, we provide the structural basis for a comprehensive description of ligand-receptor interactions that ultimately will inform new approaches to structure-based drug design.
- Paul Langerhans Institute Dresden of the Helmholtz Zentrum Munich at the University Hospital and Faculty of Medicine Carl Gustav Carus of Technische Universität Dresden, Dresden, Germany.
Organizational Affiliation: