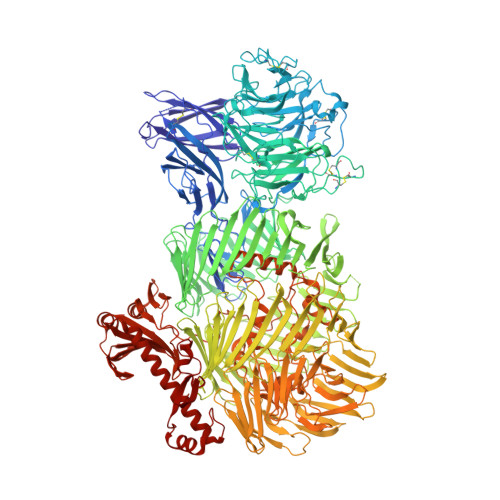
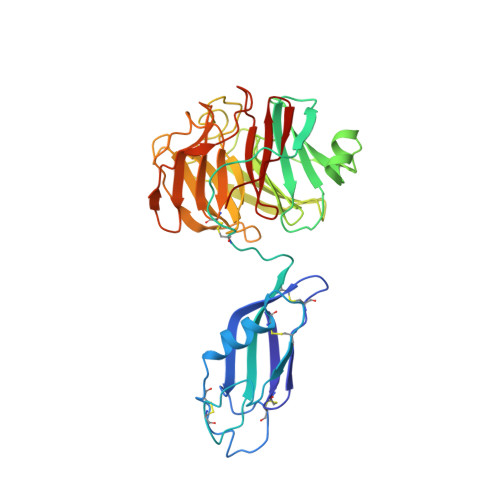
Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons.
Del Toro, D., Carrasquero-Ordaz, M.A., Chu, A., Ruff, T., Shahin, M., Jackson, V.A., Chavent, M., Berbeira-Santana, M., Seyit-Bremer, G., Brignani, S., Kaufmann, R., Lowe, E., Klein, R., Seiradake, E.(2020) Cell 180: 323-339.e19
- PubMed: 31928845
- DOI: https://doi.org/10.1016/j.cell.2019.12.014
- Primary Citation of Related Structures:
6SKA, 6SKE - PubMed Abstract:
Teneurins are ancient metazoan cell adhesion receptors that control brain development and neuronal wiring in higher animals. The extracellular C terminus binds the adhesion GPCR Latrophilin, forming a trans-cellular complex with synaptogenic functions. However, Teneurins, Latrophilins, and FLRT proteins are also expressed during murine cortical cell migration at earlier developmental stages. Here, we present crystal structures of Teneurin-Latrophilin complexes that reveal how the lectin and olfactomedin domains of Latrophilin bind across a spiraling beta-barrel domain of Teneurin, the YD shell. We couple structure-based protein engineering to biophysical analysis, cell migration assays, and in utero electroporation experiments to probe the importance of the interaction in cortical neuron migration. We show that binding of Latrophilins to Teneurins and FLRTs directs the migration of neurons using a contact repulsion-dependent mechanism. The effect is observed with cell bodies and small neurites rather than their processes. The results exemplify how a structure-encoded synaptogenic protein complex is also used for repulsive cell guidance.
- Max Planck Institute of Neurobiology, Am Klopferspitz 18, Martinsried 82152, Germany; Department of Biological Sciences, Institute of Neurosciences, IDIBAPS, CIBERNED, University of Barcelona, Barcelona, Spain.
Organizational Affiliation: