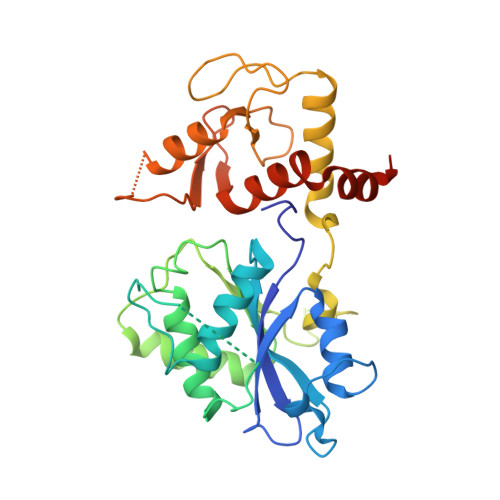
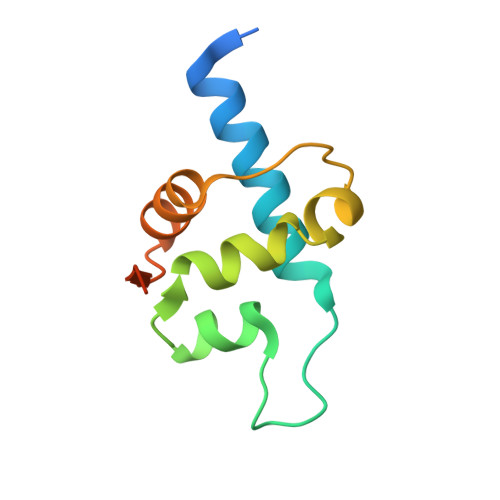
Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Gaur, V., Ziajko, W., Nirwal, S., Szlachcic, A., Gapinska, M., Nowotny, M.(2019) Nucleic Acids Res 47: 11681-11690
- PubMed: 31584081
- DOI: https://doi.org/10.1093/nar/gkz842
- Primary Citation of Related Structures:
6SEH, 6SEI - PubMed Abstract:
Structure-selective endonucleases cleave branched DNA substrates. Slx1 is unique among structure-selective nucleases because it can cleave all branched DNA structures at multiple sites near the branch point. The mechanism behind this broad range of activity is unknown. The present study structurally and biochemically investigated fungal Slx1 to define a new protein interface that binds the non-cleaved arm of branched DNAs. The DNA arm bound at this new site was positioned at a sharp angle relative to the arm that was modeled to interact with the active site, implying that Slx1 uses DNA bending to localize the branch point as a flexible discontinuity in DNA. DNA binding at the new interface promoted a disorder-order transition in a region of the protein that was located in the vicinity of the active site, potentially participating in its formation. This appears to be a safety mechanism that ensures that DNA cleavage occurs only when the new interface is occupied by the non-cleaved DNA arm. Models of Slx1 that interacted with various branched DNA substrates were prepared. These models explain the way in which Slx1 cuts DNA toward the 3' end away from the branch point and elucidate the unique ability of Slx1 to cleave various DNA structures.
- Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, 4 Trojdena St., 02-109 Warsaw, Poland.
Organizational Affiliation: