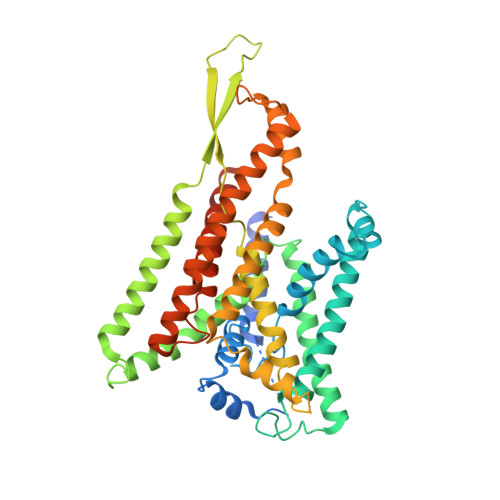
Partially inserted nascent chain unzips the lateral gate of the Sec translocon.
Kater, L., Frieg, B., Berninghausen, O., Gohlke, H., Beckmann, R., Kedrov, A.(2019) EMBO Rep 20: e48191-e48191
- PubMed: 31379073
- DOI: https://doi.org/10.15252/embr.201948191
- Primary Citation of Related Structures:
6R7L - PubMed Abstract:
The Sec translocon provides the lipid bilayer entry for ribosome-bound nascent chains and thus facilitates membrane protein biogenesis. Despite the appreciated role of the native environment in the translocon:ribosome assembly, structural information on the complex in the lipid membrane is scarce. Here, we present a cryo-electron microscopy-based structure of bacterial translocon SecYEG in lipid nanodiscs and elucidate an early intermediate state upon insertion of the FtsQ anchor domain. Insertion of the short nascent chain causes initial displacements within the lateral gate of the translocon, where α-helices 2b, 7, and 8 tilt within the membrane core to "unzip" the gate at the cytoplasmic side. Molecular dynamics simulations demonstrate that the conformational change is reversed in the absence of the ribosome, and suggest that the accessory α-helices of SecE subunit modulate the lateral gate conformation. Site-specific cross-linking validates that the FtsQ nascent chain passes the lateral gate upon insertion. The structure and the biochemical data suggest that the partially inserted nascent chain remains highly flexible until it acquires the transmembrane topology.
- Gene Center Munich, Ludwig-Maximilian-University, Munich, Germany.
Organizational Affiliation: