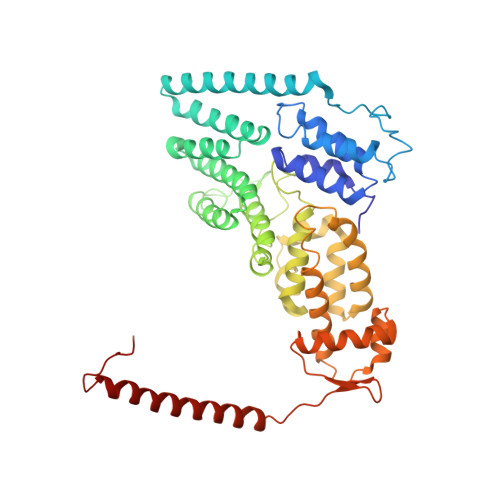
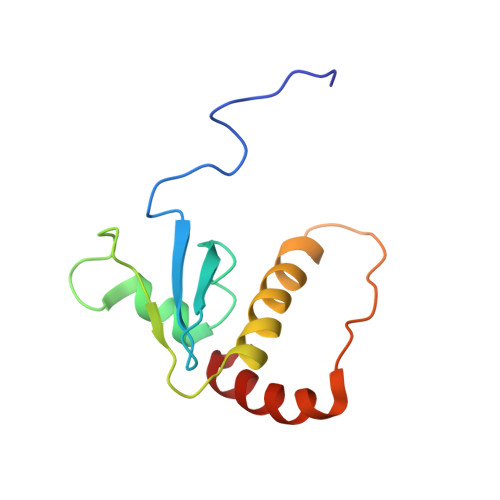
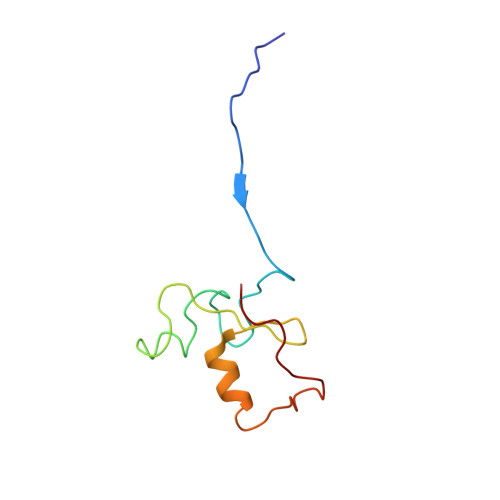
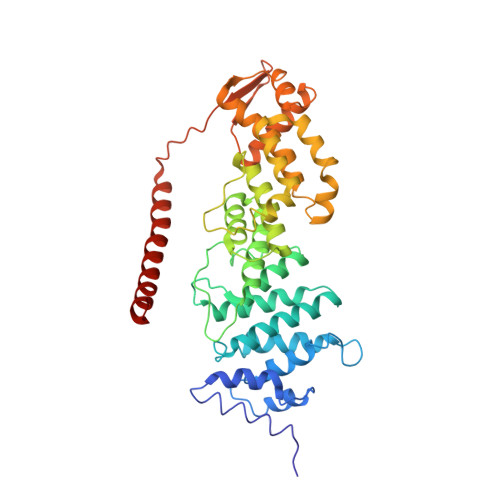
Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Faull, S.V., Lau, A.M.C., Martens, C., Ahdash, Z., Hansen, K., Yebenes, H., Schmidt, C., Beuron, F., Cronin, N.B., Morris, E.P., Politis, A.(2019) Nat Commun 10: 3814-3814
- PubMed: 31444342
- DOI: https://doi.org/10.1038/s41467-019-11772-y
- Primary Citation of Related Structures:
6R6H, 6R7F, 6R7H, 6R7I, 6R7N - PubMed Abstract:
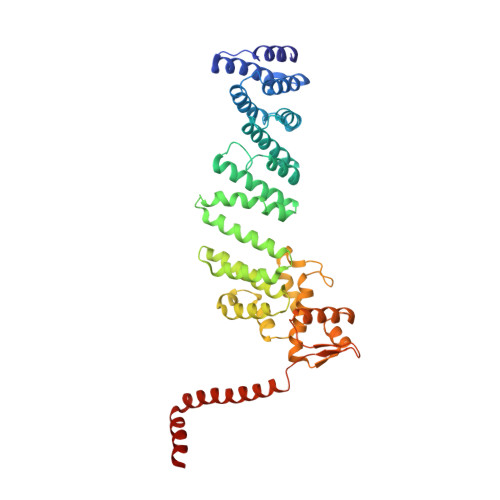
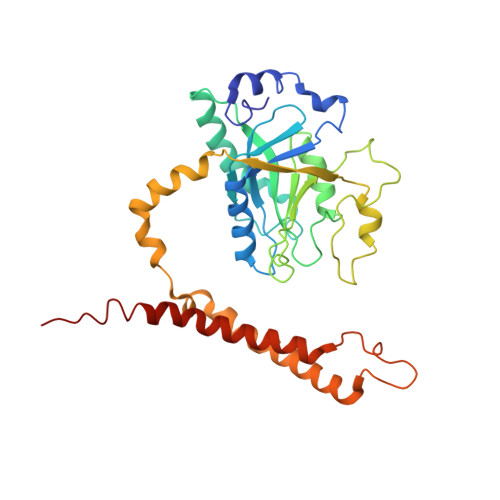
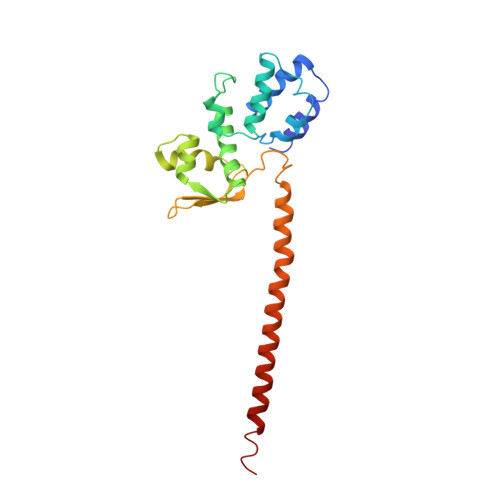
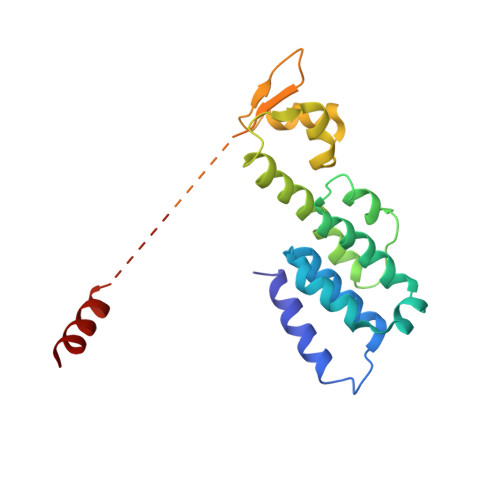
Cullin-Ring E3 Ligases (CRLs) regulate a multitude of cellular pathways through specific substrate receptors. The COP9 signalosome (CSN) deactivates CRLs by removing NEDD8 from activated Cullins. Here we present structures of the neddylated and deneddylated CSN-CRL2 complexes by combining single-particle cryo-electron microscopy (cryo-EM) with chemical cross-linking mass spectrometry (XL-MS). These structures suggest a conserved mechanism of CSN activation, consisting of conformational clamping of the CRL2 substrate by CSN2/CSN4, release of the catalytic CSN5/CSN6 heterodimer and finally activation of the CSN5 deneddylation machinery. Using hydrogen-deuterium exchange (HDX)-MS we show that CRL2 activates CSN5/CSN6 in a neddylation-independent manner. The presence of NEDD8 is required to activate the CSN5 active site. Overall, by synergising cryo-EM with MS, we identify sensory regions of the CSN that mediate its stepwise activation and provide a framework for understanding the regulatory mechanism of other Cullin family members.
- Division of Structural Biology, The Institute of Cancer Research, London, SW3 6JB, UK.
Organizational Affiliation: