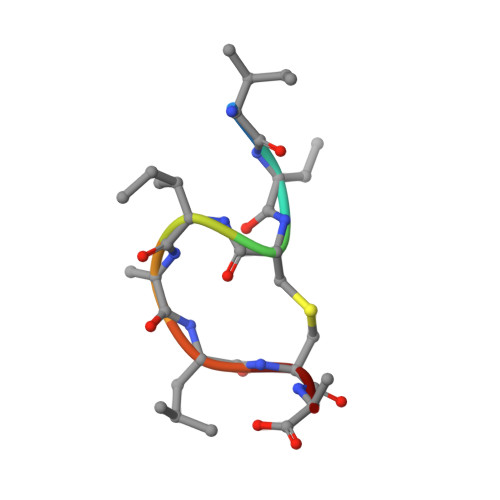
Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
Dickman, R., Mitchell, S.A., Figueiredo, A.M., Hansen, D.F., Tabor, A.B.(2019) J Org Chem 84: 11493-11512
- PubMed: 31464129
- DOI: https://doi.org/10.1021/acs.joc.9b01253
- Primary Citation of Related Structures:
6QM1, 6QTF, 6QYR, 6QYS, 6QYT, 6QYU, 6QYV, 6QYW - PubMed Abstract:
In response to the growing threat posed by antibiotic-resistant bacterial strains, extensive research is currently focused on developing antimicrobial agents that target lipid II, a vital precursor in the biosynthesis of bacterial cell walls. The lantibiotic nisin and related peptides display unique and highly selective binding to lipid II. A key feature of the nisin-lipid II interaction is the formation of a cage-like complex between the pyrophosphate moiety of lipid II and the two thioether-bridged rings, rings A and B, at the N-terminus of nisin. To understand the important structural factors underlying this highly selective molecular recognition, we have used solid-phase peptide synthesis to prepare individual ring A and B structures from nisin, the related lantibiotic mutacin, and synthetic analogues. Through NMR studies of these rings, we have demonstrated that ring A is preorganized to adopt the correct conformation for binding lipid II in solution and that individual amino acid substitutions in ring A have little effect on the conformation. We have also analyzed the turn structures adopted by these thioether-bridged peptides and show that they do not adopt the tight α-turn or β-turn structures typically found in proteins.
- Department of Chemistry , University College London , 20 Gordon Street , London WC1H 0AJ , U.K.
Organizational Affiliation: