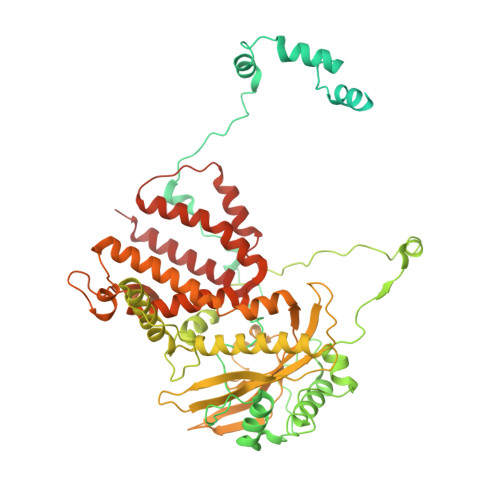
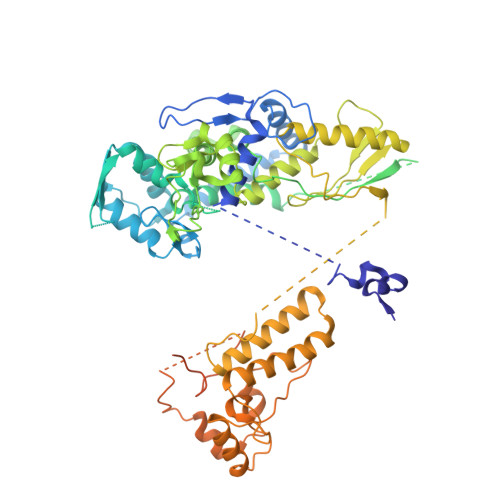
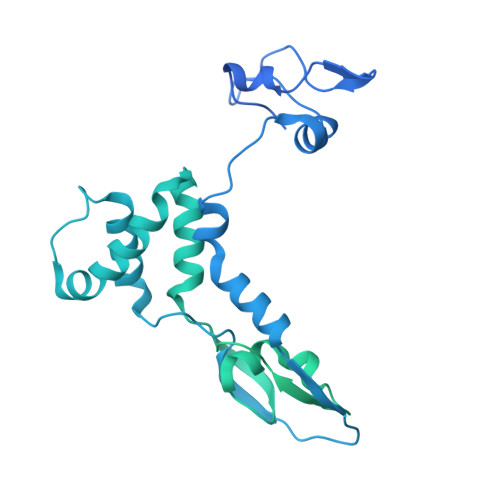
Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Fan, H., Walker, A.P., Carrique, L., Keown, J.R., Serna Martin, I., Karia, D., Sharps, J., Hengrung, N., Pardon, E., Steyaert, J., Grimes, J.M., Fodor, E.(2019) Nature 573: 287-290
- PubMed: 31485076
- DOI: https://doi.org/10.1038/s41586-019-1530-7
- Primary Citation of Related Structures:
6QNW, 6QPF, 6QPG, 6QWL, 6QX3, 6QX8, 6QXE, 6RR7 - PubMed Abstract:
Influenza A viruses are responsible for seasonal epidemics, and pandemics can arise from the transmission of novel zoonotic influenza A viruses to humans 1,2 . Influenza A viruses contain a segmented negative-sense RNA genome, which is transcribed and replicated by the viral-RNA-dependent RNA polymerase (FluPol A ) composed of PB1, PB2 and PA subunits 3-5 . Although the high-resolution crystal structure of FluPol A of bat influenza A virus has previously been reported 6 , there are no complete structures available for human and avian FluPol A . Furthermore, the molecular mechanisms of genomic viral RNA (vRNA) replication-which proceeds through a complementary RNA (cRNA) replicative intermediate, and requires oligomerization of the polymerase 7-10 -remain largely unknown. Here, using crystallography and cryo-electron microscopy, we determine the structures of FluPol A from human influenza A/NT/60/1968 (H3N2) and avian influenza A/duck/Fujian/01/2002 (H5N1) viruses at a resolution of 3.0-4.3 Å, in the presence or absence of a cRNA or vRNA template. In solution, FluPol A forms dimers of heterotrimers through the C-terminal domain of the PA subunit, the thumb subdomain of PB1 and the N1 subdomain of PB2. The cryo-electron microscopy structure of monomeric FluPol A bound to the cRNA template reveals a binding site for the 3' cRNA at the dimer interface. We use a combination of cell-based and in vitro assays to show that the interface of the FluPol A dimer is required for vRNA synthesis during replication of the viral genome. We also show that a nanobody (a single-domain antibody) that interferes with FluPol A dimerization inhibits the synthesis of vRNA and, consequently, inhibits virus replication in infected cells. Our study provides high-resolution structures of medically relevant FluPol A , as well as insights into the replication mechanisms of the viral RNA genome. In addition, our work identifies sites in FluPol A that could be targeted in the development of antiviral drugs.
- Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.
Organizational Affiliation: