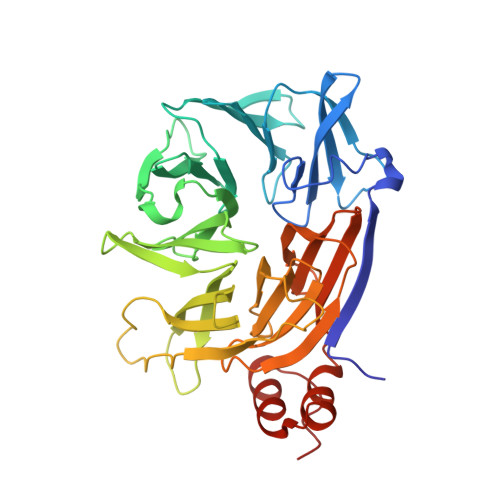
Clathrin's adaptor interaction sites are repurposed to stabilize microtubules during mitosis.
Rondelet, A., Lin, Y.C., Singh, D., Porfetye, A.T., Thakur, H.C., Hecker, A., Brinkert, P., Schmidt, N., Bendre, S., Muller, F., Mazul, L., Widlund, P.O., Bange, T., Hiller, M., Vetter, I.R., Bird, A.W.(2020) J Cell Biol 219
- PubMed: 31932847
- DOI: https://doi.org/10.1083/jcb.201907083
- Primary Citation of Related Structures:
6QNN, 6QNP - PubMed Abstract:
Clathrin ensures mitotic spindle stability and efficient chromosome alignment, independently of its vesicle trafficking function. Although clathrin localizes to the mitotic spindle and kinetochore fiber microtubule bundles, the mechanisms by which clathrin stabilizes microtubules are unclear. We show that clathrin adaptor interaction sites on clathrin heavy chain (CHC) are repurposed during mitosis to directly recruit the microtubule-stabilizing protein GTSE1 to the spindle. Structural analyses reveal that these sites interact directly with clathrin-box motifs on GTSE1. Disruption of this interaction releases GTSE1 from spindles, causing defects in chromosome alignment. Surprisingly, this disruption destabilizes astral microtubules, but not kinetochore-microtubule attachments, and chromosome alignment defects are due to a failure of chromosome congression independent of kinetochore-microtubule attachment stability. GTSE1 recruited to the spindle by clathrin stabilizes microtubules by inhibiting the microtubule depolymerase MCAK. This work uncovers a novel role of clathrin adaptor-type interactions to stabilize nonkinetochore fiber microtubules to support chromosome congression, defining for the first time a repurposing of this endocytic interaction mechanism during mitosis.
- Max Planck Institute of Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: