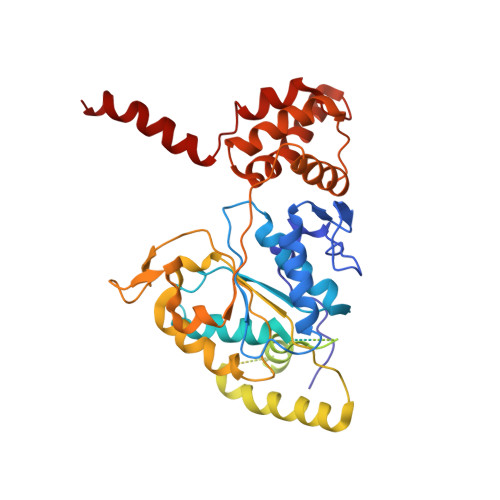
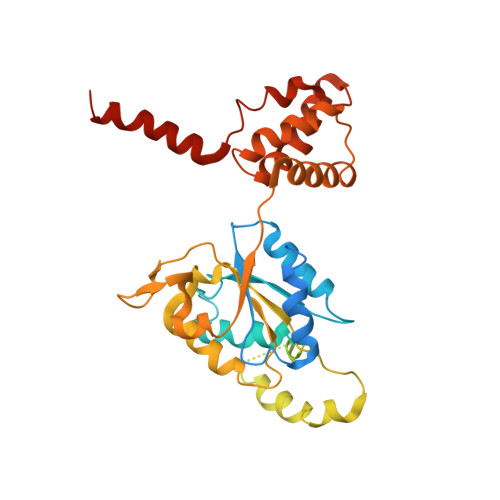
Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Munoz-Hernandez, H., Pal, M., Rodriguez, C.F., Fernandez-Leiro, R., Prodromou, C., Pearl, L.H., Llorca, O.(2019) Sci Adv 5: eaaw1616-eaaw1616
- PubMed: 31049401
- DOI: https://doi.org/10.1126/sciadv.aaw1616
- Primary Citation of Related Structures:
6QI8, 6QI9 - PubMed Abstract:
The human R2TP complex (RUVBL1-RUVBL2-RPAP3-PIH1D1) is an HSP90 co-chaperone required for the maturation of several essential multiprotein complexes, including RNA polymerase II, small nucleolar ribonucleoproteins, and PIKK complexes such as mTORC1 and ATR-ATRIP. RUVBL1-RUVBL2 AAA-ATPases are also primary components of other essential complexes such as INO80 and Tip60 remodelers. Despite recent efforts, the molecular mechanisms regulating RUVBL1-RUVBL2 in these complexes remain elusive. Here, we report cryo-EM structures of R2TP and show how access to the nucleotide-binding site of RUVBL2 is coupled to binding of the client recruitment component of R2TP (PIH1D1) to its DII domain. This interaction induces conformational rearrangements that lead to the destabilization of an N-terminal segment of RUVBL2 that acts as a gatekeeper to nucleotide exchange. This mechanism couples protein-induced motions of the DII domains with accessibility of the nucleotide-binding site in RUVBL1-RUVBL2, and it is likely a general mechanism shared with other RUVBL1-RUVBL2-containing complexes.
- Structural Biology Programme, Spanish National Cancer Research Centre (CNIO), Calle de Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Organizational Affiliation: