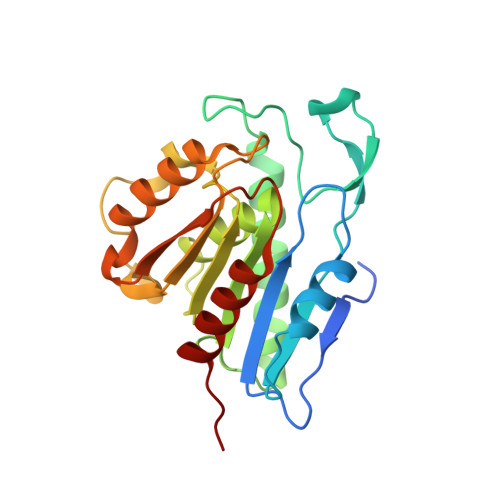
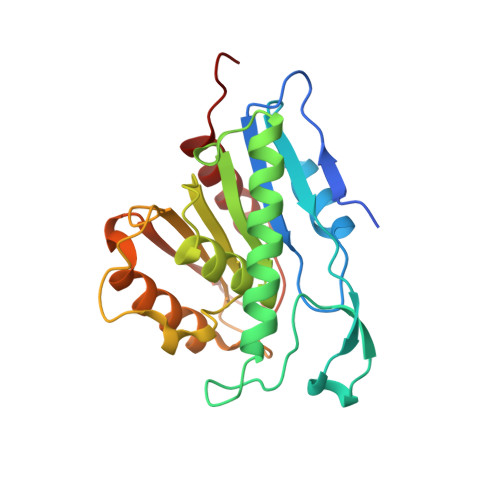
Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Abrami, L., Audagnotto, M., Ho, S., Marcaida, M.J., Mesquita, F.S., Anwar, M.U., Sandoz, P.A., Fonti, G., Pojer, F., Dal Peraro, M., van der Goot, F.G.(2021) Nat Chem Biol
- PubMed: 33707782
- DOI: https://doi.org/10.1038/s41589-021-00753-2
- Primary Citation of Related Structures:
6QGN, 6QGO, 6QGQ, 6QGS - PubMed Abstract:
Many biochemical reactions require controlled recruitment of proteins to membranes. This is largely regulated by posttranslational modifications. A frequent one is S-acylation, which consists of the addition of acyl chains and can be reversed by poorly understood acyl protein thioesterases (APTs). Using a panel of computational and experimental approaches, we dissect the mode of action of the major cellular thioesterase APT2 (LYPLA2). We show that soluble APT2 is vulnerable to proteasomal degradation, from which membrane binding protects it. Interaction with membranes requires three consecutive steps: electrostatic attraction, insertion of a hydrophobic loop and S-acylation by the palmitoyltransferases ZDHHC3 or ZDHHC7. Once bound, APT2 is predicted to deform the lipid bilayer to extract the acyl chain bound to its substrate and capture it in a hydrophobic pocket to allow hydrolysis. This molecular understanding of APT2 paves the way to understand the dynamics of APT2-mediated deacylation of substrates throughout the endomembrane system.
- Global Health Institute, School of Life Sciences, EPFL, Lausanne, Switzerland.
Organizational Affiliation: