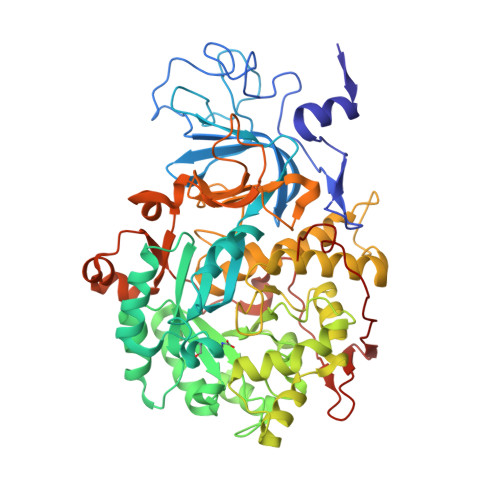
The Structure of the Elusive Urease-Urea Complex Unveils the Mechanism of a Paradigmatic Nickel-Dependent Enzyme.
Mazzei, L., Cianci, M., Benini, S., Ciurli, S.(2019) Angew Chem Int Ed Engl 58: 7415-7419
- PubMed: 30969470
- DOI: https://doi.org/10.1002/anie.201903565
- Primary Citation of Related Structures:
6QDY - PubMed Abstract:
Urease, the most efficient enzyme known, contains an essential dinuclear Ni II cluster in the active site. It catalyzes the hydrolysis of urea, inducing a rapid pH increase that has negative effects on human health and agriculture. Thus, the control of urease activity is of utmost importance in medical, pharmaceutical, and agro-environmental applications. All known urease inhibitors are either toxic or inefficient. The development of new and efficient chemicals able to inhibit urease relies on the knowledge of all steps of the catalytic mechanism. The short (microseconds) lifetime of the urease-urea complex has hampered the determination of its structure. The present study uses fluoride to substitute the hydroxide acting as the co-substrate in the reaction, preventing the occurrence of the catalytic steps that follow substrate binding. The 1.42 Å crystal structure of the urease-urea complex, reported here, resolves the enduring debate on the mechanism of this metalloenzyme.
- Laboratory of Bioinorganic Chemistry, Departement of Pharmacy and Biotechnology, University of Bologna, Via Giuseppe Fanin 40, 40138, Bologna, Italy.
Organizational Affiliation: