Cryo-EM structure of Lysine decarboxylase A from Pseudomonas aeruginosa
Kandiah, E., Gutsche, I.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Biodegradative arginine decarboxylase | 751 | Pseudomonas aeruginosa | Mutation(s): 0 Gene Names: adiA, adiA_2, CAZ10_25795, CGU42_31920, DY979_03865, DZ962_05275, EB236_18185, EGV95_19425, EGY23_25530, IPC3_20945... EC: 4.1.1.19 (PDB Primary Data), 4.1.1.18 (PDB Primary Data) | 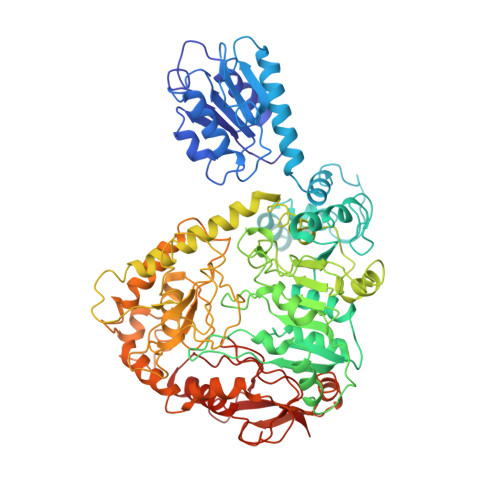 | |
UniProt | |||||
Find proteins for Q9I2S7 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore Q9I2S7 Go to UniProtKB: Q9I2S7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9I2S7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PLP Query on PLP | B [auth A] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 1.4 |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| French National Research Agency | France | ANR-12-JSV8-0002 |
| European Research Council | France | ERC 647784 |