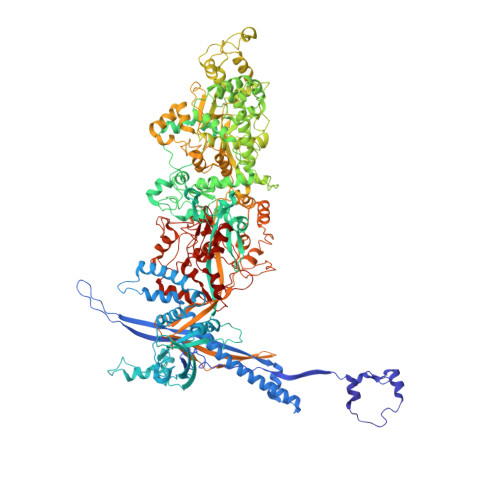
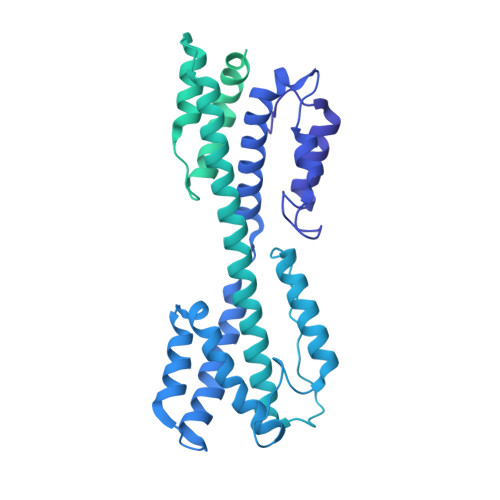
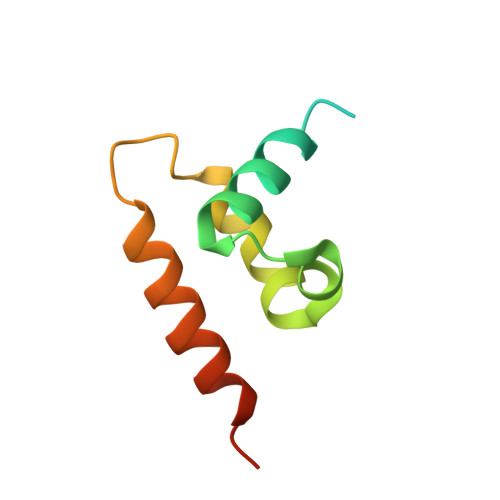
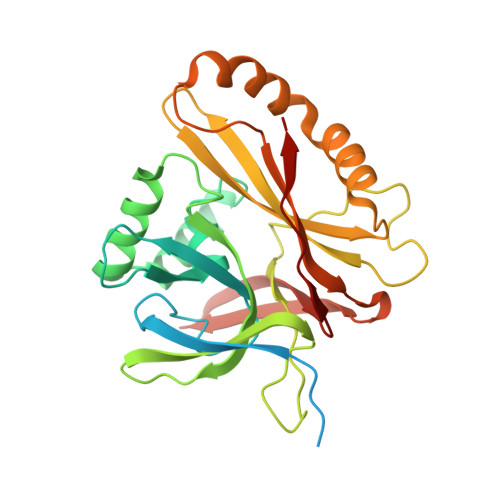
Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes.
Zhang, Y., Liu, W., Li, Z., Kumar, V., Alvarez-Cabrera, A.L., Leibovitch, E.C., Cui, Y., Mei, Y., Bi, G.Q., Jacobson, S., Zhou, Z.H.(2019) Nat Commun 10: 5346-5346
- PubMed: 31767868
- DOI: https://doi.org/10.1038/s41467-019-13064-x
- Primary Citation of Related Structures:
6Q1F - PubMed Abstract:
Human herpesvirus 6B (HHV-6B) belongs to the β-herpesvirus subfamily of the Herpesviridae. To understand capsid assembly and capsid-tegument interactions, here we report atomic structures of HHV-6B capsid and capsid-associated tegument complex (CATC) obtained by cryoEM and sub-particle reconstruction. Compared to other β-herpesviruses, HHV-6B exhibits high similarity in capsid structure but organizational differences in its CATC (pU11 tetramer). 180 "VΛ"-shaped CATCs are observed in HHV-6B, distinguishing from the 255 "Λ"-shaped dimeric CATCs observed in murine cytomegalovirus and the 310 "Δ"-shaped CATCs in human cytomegalovirus. This trend in CATC quantity correlates with the increasing genomes sizes of these β-herpesviruses. Incompatible distances revealed by the atomic structures rationalize the lack of CATC's binding to triplexes Ta, Tc, and Tf in HHV-6B. Our results offer insights into HHV-6B capsid assembly and the roles of its tegument proteins, including not only the β-herpesvirus-specific pU11 and pU14, but also those conserved across all subfamilies of Herpesviridae.
- Center for Integrative Imaging, Hefei National Laboratory for Physical Sciences at the Microscale, and School of Life Sciences, University of Science and Technology of China (USTC), Hefei, Anhui, 230026, China.
Organizational Affiliation: