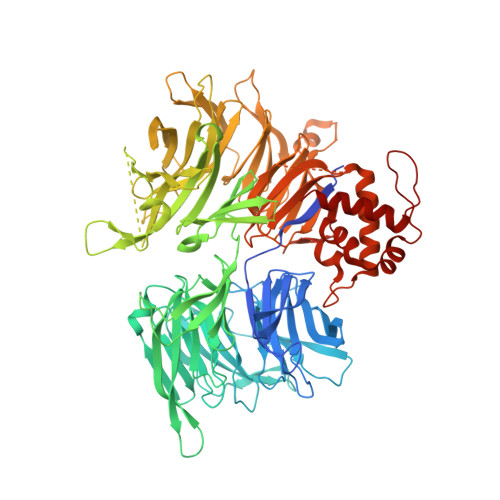
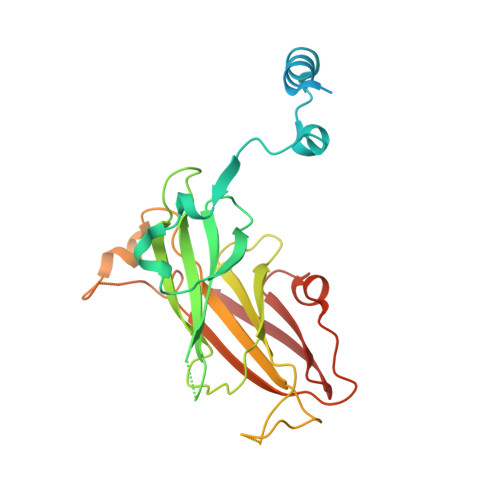
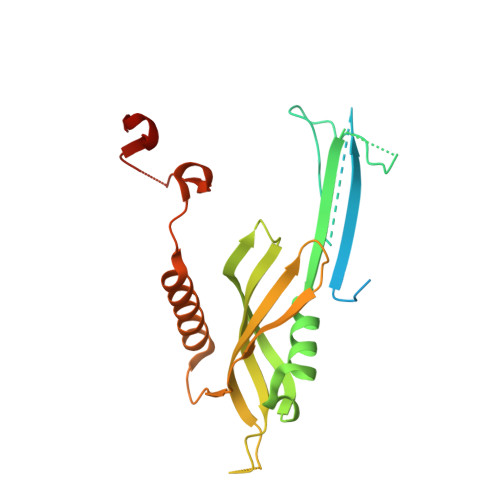
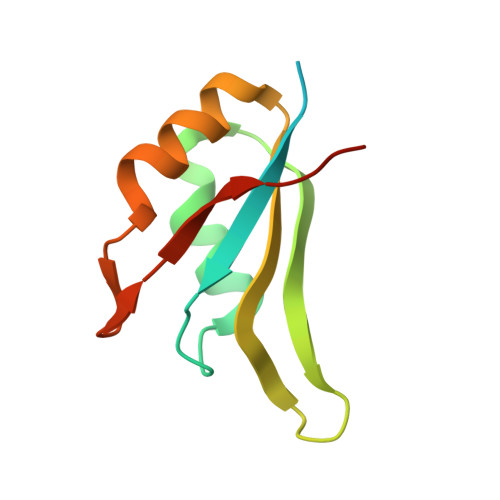
Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Faust, T.B., Yoon, H., Nowak, R.P., Donovan, K.A., Li, Z., Cai, Q., Eleuteri, N.A., Zhang, T., Gray, N.S., Fischer, E.S.(2020) Nat Chem Biol 16: 7-14
- PubMed: 31686031
- DOI: https://doi.org/10.1038/s41589-019-0378-3
- Primary Citation of Related Structures:
6Q0R, 6Q0V, 6Q0W - PubMed Abstract:
The investigational drugs E7820, indisulam and tasisulam (aryl-sulfonamides) promote the degradation of the splicing factor RBM39 in a proteasome-dependent mechanism. While the activity critically depends on the cullin RING ligase substrate receptor DCAF15, the molecular details remain elusive. Here we present the cryo-EM structure of the DDB1-DCAF15-DDA1 core ligase complex bound to RBM39 and E7820 at a resolution of 4.4 Å, together with crystal structures of engineered subcomplexes. We show that DCAF15 adopts a new fold stabilized by DDA1, and that extensive protein-protein contacts between the ligase and substrate mitigate low affinity interactions between aryl-sulfonamides and DCAF15. Our data demonstrate how aryl-sulfonamides neo-functionalize a shallow, non-conserved pocket on DCAF15 to selectively bind and degrade RBM39 and the closely related splicing factor RBM23 without the requirement for a high-affinity ligand, which has broad implications for the de novo discovery of molecular glue degraders.
- Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, USA.
Organizational Affiliation: