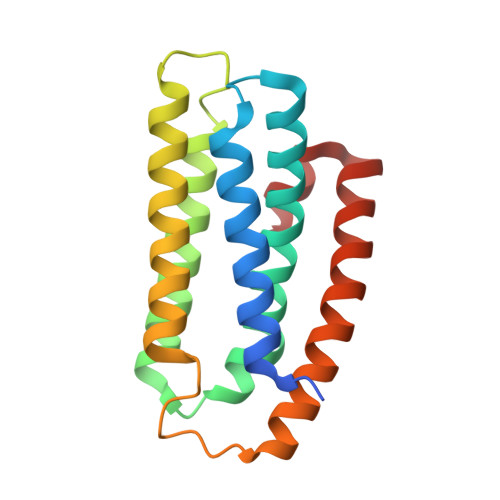
Crystal structure of a hemerythrin-like protein from Mycobacterium kansasii and homology model of the orthologous Rv2633c protein of M. tuberculosis.
Ma, Z., Abendroth, J., Buchko, G.W., Rohde, K.H., Davidson, V.L.(2020) Biochem J 477: 567-581
- PubMed: 31913442
- DOI: https://doi.org/10.1042/BCJ20190827
- Primary Citation of Related Structures:
6Q09, 6U3L - PubMed Abstract:
Pathogenic and opportunistic mycobacteria have a distinct class of non-heme di-iron hemerythrin-like proteins (HLPs). The first to be isolated was the Rv2633c protein, which plays a role in infection by Mycobacterium tuberculosis (Mtb), but could not be crystallized. This work presents the first crystal structure of an ortholog of Rv2633c, the mycobacterial HLP from Mycobacterium kansasii (Mka). This structure differs from those of hemerythrins and other known HLPs. It consists of five α-helices, whereas all other HLP domains have four. In contrast with other HLPs, the HLP domain is not fused to an additional protein domain. The residues ligating and surrounding the di-iron site are also unique among HLPs. Notably, a tyrosine occupies the position normally held by one of the histidine ligands in hemerythrin. This structure was used to construct a homology model of Rv2633c. The structure of five α-helices is conserved and the di-iron site ligands are identical in Rv2633c. Two residues near the ends of helices in the Mka HLP structure are replaced with prolines in the Rv2633c model. This may account for structural perturbations that decrease the solubility of Rv2633c relative to Mka HLP. Clusters of residues that differ in charge or polarity between Rv2633c and Mka HLP that point outward from the helical core could reflect a specificity for potential differential interactions with other protein partners in vivo, which are related to function. The Mka HLP exhibited weaker catalase activity than Rv2633c. Evidence was obtained for the interaction of Mka HLP irons with nitric oxide.
- Burnett School of Biomedical Sciences, College of Medicine, University of Central Florida, Orlando, FL 32827, U.S.A.
Organizational Affiliation: