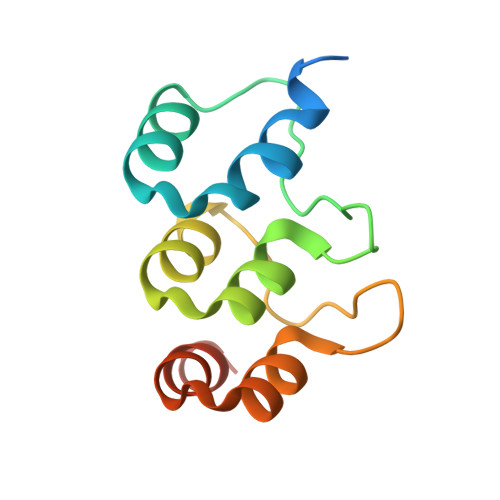
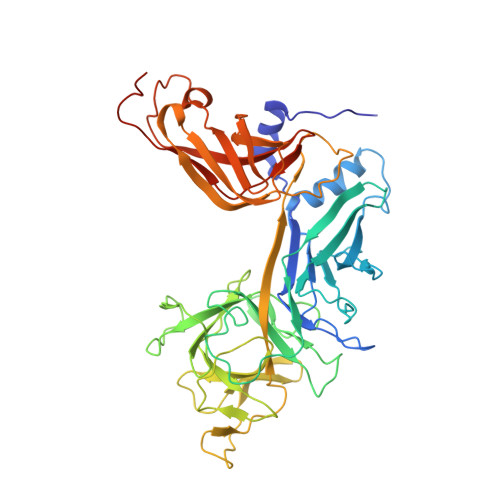
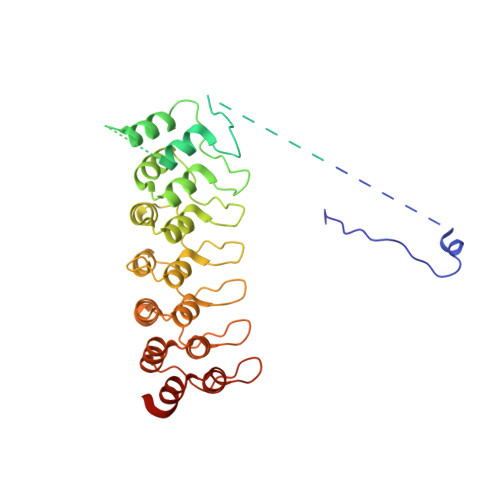
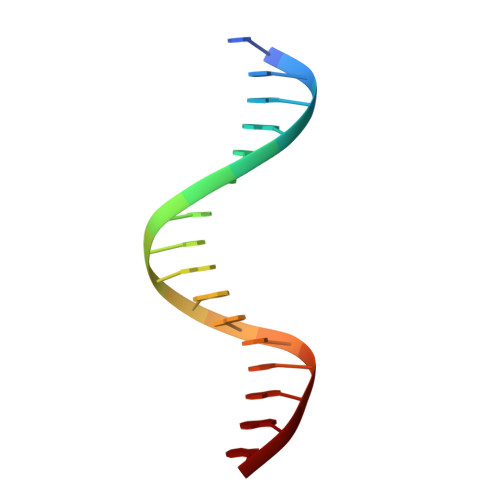
Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Jarrett, S.M., Seegar, T.C.M., Andrews, M., Adelmant, G., Marto, J.A., Aster, J.C., Blacklow, S.C.(2019) Sci Signal 12
- PubMed: 31690634
- DOI: https://doi.org/10.1126/scisignal.aay2369
- Primary Citation of Related Structures:
6PY8 - PubMed Abstract:
Canonical Notch signaling relies on regulated proteolysis of the receptor Notch to generate a nuclear effector that induces the transcription of Notch-responsive genes. In higher organisms, one Notch-responsive gene that is activated in many different cell types encodes the Notch-regulated ankyrin repeat protein (NRARP), which acts as a negative feedback regulator of Notch responses. Here, we showed that NRARP inhibited the growth of Notch-dependent T cell acute lymphoblastic leukemia (T-ALL) cell lines and bound directly to the core Notch transcriptional activation complex (NTC), requiring both the transcription factor RBPJ and the Notch intracellular domain (NICD), but not Mastermind-like proteins or DNA. The crystal structure of an NRARP-NICD1-RBPJ-DNA complex, determined to 3.75 Å resolution, revealed that the assembly of NRARP-NICD1-RBPJ complexes relied on simultaneous engagement of RBPJ and NICD1, with the three ankyrin repeats of NRARP extending the Notch1 ankyrin repeat stack. Mutations at the NRARP-NICD1 interface disrupted entry of the proteins into NTCs and abrogated feedback inhibition in Notch signaling assays in cultured cells. Forced expression of NRARP reduced the abundance of NICD in cells, suggesting that NRARP may promote the degradation of NICD. These studies establish the structural basis for NTC engagement by NRARP and provide insights into a critical negative feedback mechanism that regulates Notch signaling.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: