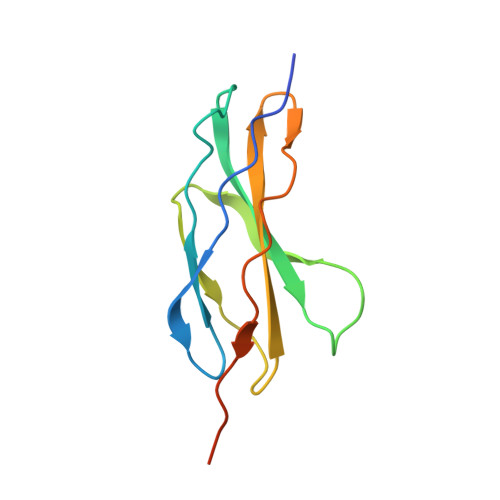
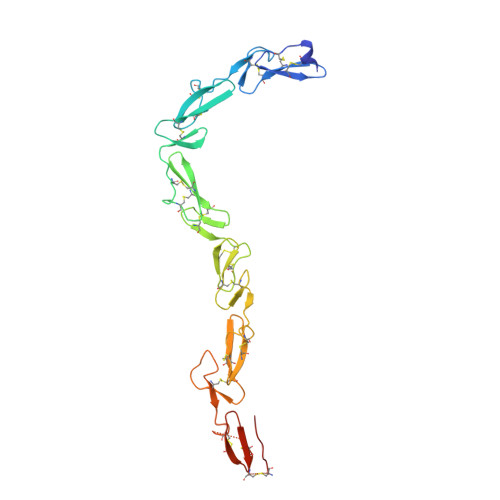
NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Pak, J.S., DeLoughery, Z.J., Wang, J., Acharya, N., Park, Y., Jaworski, A., Ozkan, E.(2020) Nat Commun 11: 1489-1489
- PubMed: 32198364
- DOI: https://doi.org/10.1038/s41467-020-15211-1
- Primary Citation of Related Structures:
6POG, 6POK, 6POL - PubMed Abstract:
Axon pathfinding is critical for nervous system development, and it is orchestrated by molecular cues that activate receptors on the axonal growth cone. Robo family receptors bind Slit guidance cues to mediate axon repulsion. In mammals, the divergent family member Robo3 does not bind Slits, but instead signals axon repulsion from its own ligand, NELL2. Conversely, canonical Robos do not mediate NELL2 signaling. Here, we present the structures of NELL-Robo3 complexes, identifying a mode of ligand engagement for Robos that is orthogonal to Slit binding. We elucidate the structural basis for differential binding between NELL and Robo family members and show that NELL2 repulsive activity is a function of its Robo3 affinity and is enhanced by ligand trimerization. Our results reveal a mechanism of oligomerization-induced Robo activation for axon guidance and shed light on Robo family member ligand binding specificity, conformational variability, divergent modes of signaling, and evolution.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL, 60637, USA.
Organizational Affiliation: