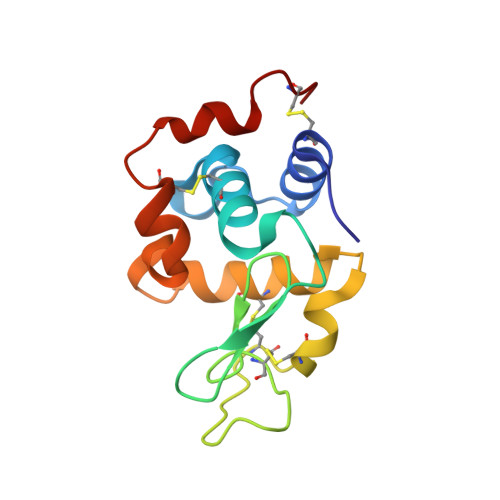
Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Truong, J.Q., Panjikar, S., Shearwin-Whyatt, L., Bruning, J.B., Shearwin, K.E.(2019) Acta Crystallogr D Struct Biol 75: 670-681
- PubMed: 31282476
- DOI: https://doi.org/10.1107/S2059798319009008
- Primary Citation of Related Structures:
6O43, 6PBB - PubMed Abstract:
Two commonly encountered bottlenecks in the structure determination of a protein by X-ray crystallography are screening for conditions that give high-quality crystals and, in the case of novel structures, finding derivatization conditions for experimental phasing. In this study, the phasing molecule 5-amino-2,4,6-triiodoisophthalic acid (I3C) was added to a random microseed matrix screen to generate high-quality crystals derivatized with I3C in a single optimization experiment. I3C, often referred to as the magic triangle, contains an aromatic ring scaffold with three bound I atoms. This approach was applied to efficiently phase the structures of hen egg-white lysozyme and the N-terminal domain of the Orf11 protein from Staphylococcus phage P68 (Orf11 NTD) using SAD phasing. The structure of Orf11 NTD suggests that it may play a role as a virion-associated lysin or endolysin.
- School of Biological Sciences, The University of Adelaide, North Terrace, Adelaide, South Australia 5005, Australia.
Organizational Affiliation: