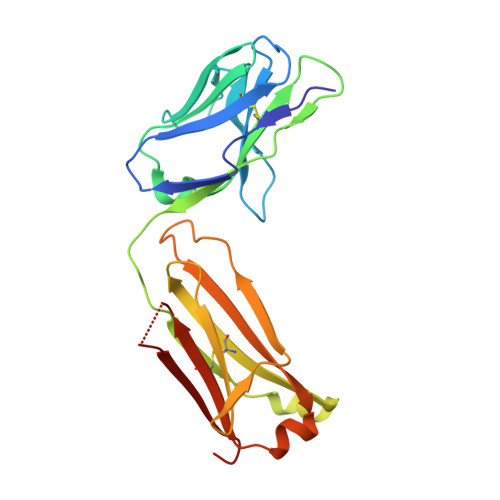
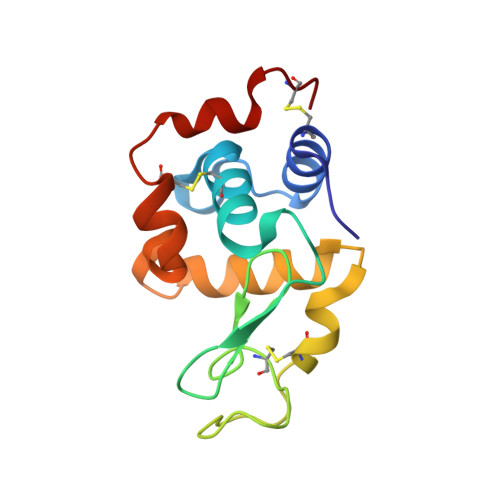
Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Burnett, D.L., Schofield, P., Langley, D.B., Jackson, J., Bourne, K., Wilson, E., Porebski, B.T., Buckle, A.M., Brink, R., Goodnow, C.C., Christ, D.(2020) Proc Natl Acad Sci U S A 117: 22341-22350
- PubMed: 32855302
- DOI: https://doi.org/10.1073/pnas.2005102117
- Primary Citation of Related Structures:
6P4A, 6P4B, 6P4C, 6P4D - PubMed Abstract:
Conformational diversity and self-cross-reactivity of antigens have been correlated with evasion from neutralizing antibody responses. We utilized single cell B cell sequencing, biolayer interferometry and X-ray crystallography to trace mutation selection pathways where the antibody response must resolve cross-reactivity between foreign and self-proteins bearing near-identical contact surfaces, but differing in conformational flexibility. Recurring antibody mutation trajectories mediate long-range rearrangements of framework (FW) and complementarity determining regions (CDRs) that increase binding site conformational diversity. These antibody mutations decrease affinity for self-antigen 19-fold and increase foreign affinity 67-fold, to yield a more than 1,250-fold increase in binding discrimination. These results demonstrate how conformational diversity in antigen and antibody does not act as a barrier, as previously suggested, but rather facilitates high affinity and high discrimination between foreign and self.
- Garvan Institute of Medical Research, University of New South Wales (UNSW) Sydney, Darlinghurst, NSW 2010, Australia.
Organizational Affiliation: