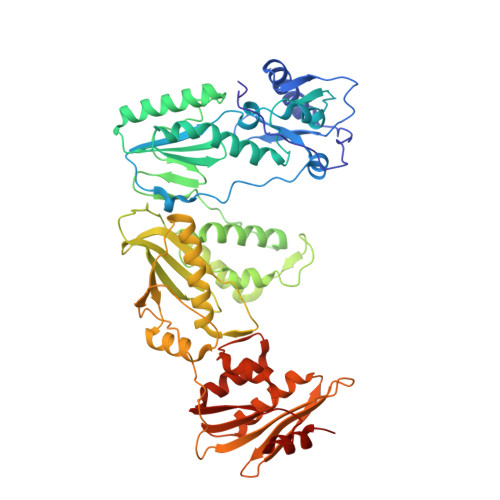
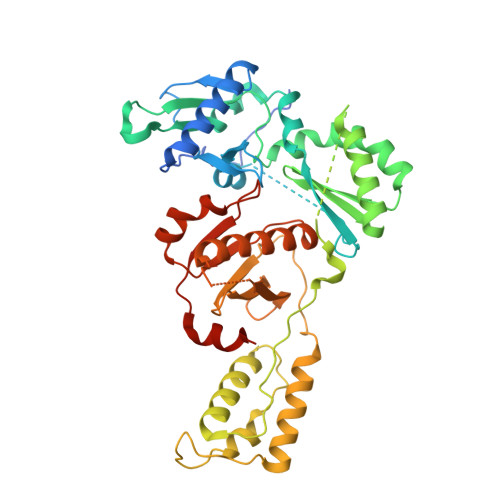
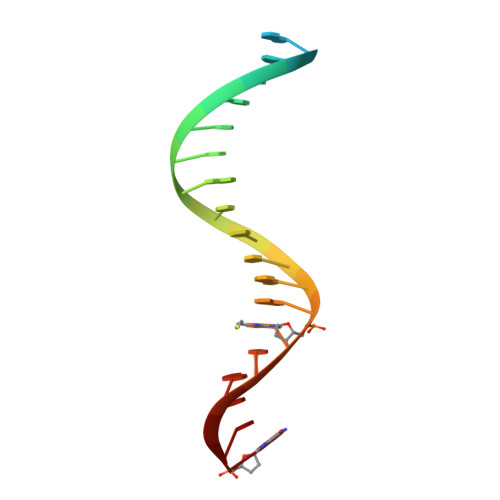
Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Bertoletti, N., Chan, A.H., Schinazi, R.F., Yin, Y.W., Anderson, K.S.(2019) Protein Sci 28: 1664-1675
- PubMed: 31301259
- DOI: https://doi.org/10.1002/pro.3681
- Primary Citation of Related Structures:
6OR7, 6OTZ, 6OUN, 6P1I, 6P1X, 6P2G - PubMed Abstract:
The retrovirus HIV-1 has been a major health issue since its discovery in the early 80s. In 2017, over 37 million people were infected with HIV-1, of which 1.8 million were new infections that year. Currently, the most successful treatment regimen is the highly active antiretroviral therapy (HAART), which consists of a combination of three to four of the current 26 FDA-approved HIV-1 drugs. Half of these drugs target the reverse transcriptase (RT) enzyme that is essential for viral replication. One class of RT inhibitors is nucleoside reverse transcriptase inhibitors (NRTIs), a crucial component of the HAART. Once incorporated into DNA, NRTIs function as a chain terminator to stop viral DNA replication. Unfortunately, treatment with NRTIs is sometimes linked to toxicity caused by off-target side effects. NRTIs may also target the replicative human mitochondrial DNA polymerase (Pol γ), causing long-term severe drug toxicity. The goal of this work is to understand the discrimination mechanism of different NRTI analogues by RT. Crystal structures and kinetic experiments are essential for the rational design of new molecules that are able to bind selectively to RT and not Pol γ. Structural comparison of NRTI-binding modes with both RT and Pol γ enzymes highlights key amino acids that are responsible for the difference in affinity of these drugs to their targets. Therefore, the long-term goal of this research is to develop safer, next generation therapeutics that can overcome off-target toxicity.
- Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut.
Organizational Affiliation: