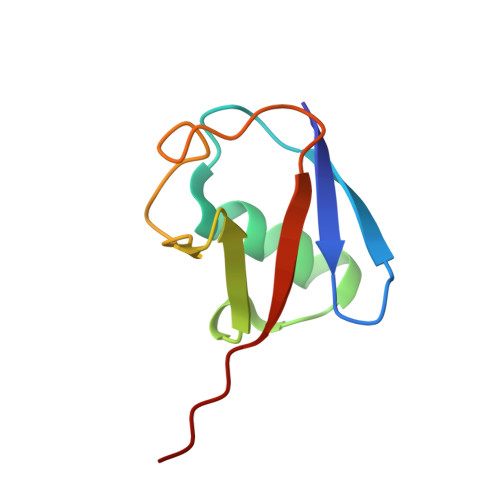
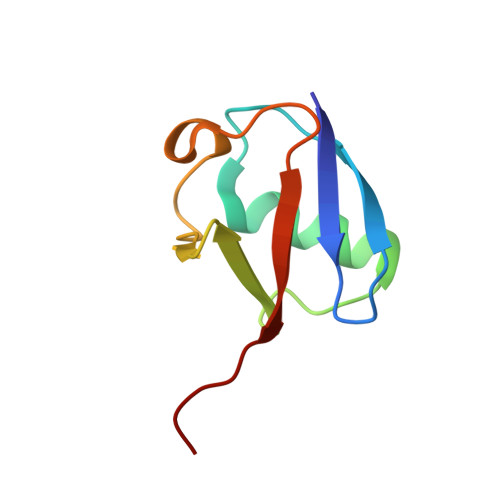
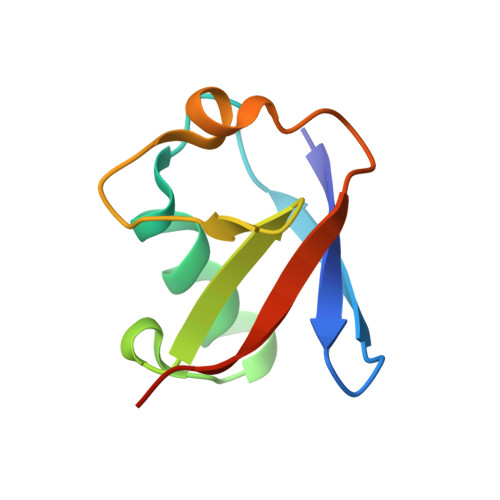
Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1.
Boughton, A.J., Krueger, S., Fushman, D.(2020) Structure 28: 29-43.e6
- PubMed: 31677892
- DOI: https://doi.org/10.1016/j.str.2019.10.008
- Primary Citation of Related Structures:
6OQ1, 6OQ2 - PubMed Abstract:
Post-translational substrate modification with ubiquitin is essential for eukaryotic cellular signaling. Polymeric ubiquitin chains are assembled with specific architectures, which convey distinct signaling outcomes depending on the linkages involved. Recently, branched K11/K48-linked polyubiquitins were shown to enhance proteasomal degradation during mitosis. To better understand the underlying structural mechanisms, we determined the crystal and NMR structures of branched K11/K48-linked tri-ubiquitin and discovered a previously unobserved interdomain interface between the distal ubiquitins. Small-angle neutron scattering and site-directed mutagenesis corroborated the presence of this interface, which we hypothesized to be influential in the physiological role of branched K11/K48-linked chains. Yet, experiments probing polyubiquitin interactions-deubiquitination assays, binding to proteasomal shuttle hHR23A-showed negligible differences between branched K11/K48-linked tri-ubiquitin and related di-ubiquitins. However, significantly stronger binding affinity for branched K11/K48-linked tri-ubiquitin was observed with proteasomal subunit Rpn1, thereby suggesting a functional impact of this interdomain interface and pinpointing the mechanistic site of enhanced degradation.
- Department of Chemistry and Biochemistry, Center for Biomolecular Structure and Organization, University of Maryland, College Park, MD 20742, USA.
Organizational Affiliation: