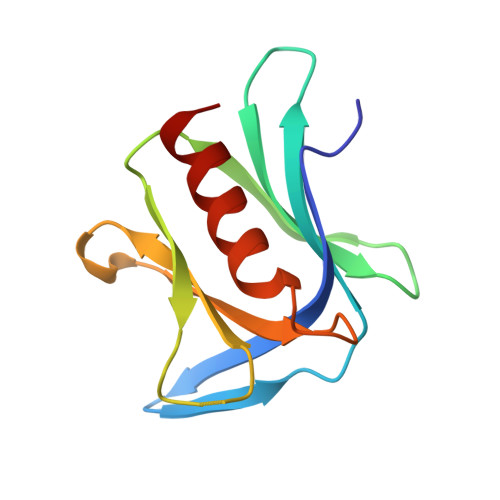
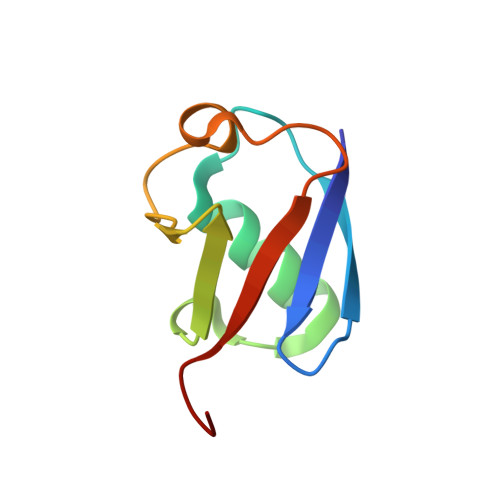
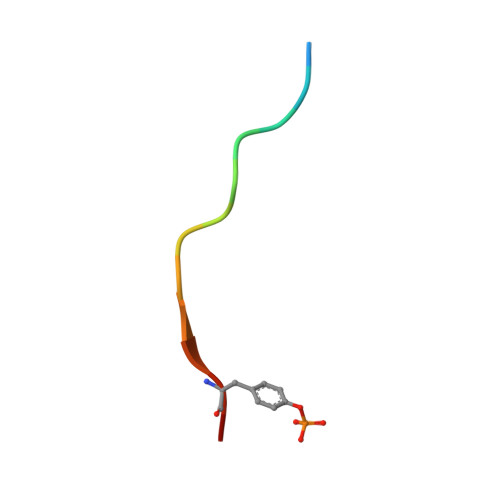
Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
Hemmis, C.W., Heard, S.C., Hill, C.P.(2019) J Biological Chem 294: 9659-9665
- PubMed: 31064842
- DOI: https://doi.org/10.1074/jbc.AC119.008881
- Primary Citation of Related Structures:
6OI4 - PubMed Abstract:
Protein substrates are targeted to the 26S proteasome through several ubiquitin receptors. One of these receptors, RPN13, is recruited to the proteasome by binding of its N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain to C-terminal residues of the scaffolding protein RPN2. The RPN13 PRU domain is followed by a flexible linker and a C-terminal deubiquitylase adaptor (DEUBAD) domain, which recruits and activates the deubiquitylase UCH37. Both RPN13 and UCH37 have been implicated in human cancers, and inhibitors of the RPN2-RPN13 interaction are being developed as potential therapeutic anticancer agents. Our current study builds on the recognition that a residue central to the RPN2-RPN13 interaction, RPN2 Tyr-950, is phosphorylated in Jurkat cells. We found that the Tyr-950 phosphorylation enhances binding to RPN13. The crystal structure of the RPN2-RPN13 pTyr-950-ubiquitin complex was determined at 1.76-Å resolution and reveals specific interactions with positively charged side chains in RPN13 that explain how phosphorylation increases binding affinity without inducing conformational change. Mutagenesis and quantitative binding assays were then used to validate the crystallographic interface. Our findings support a model in which RPN13 recruitment to the proteasome is enhanced by phosphorylation of RPN2 Tyr-950, have important implications for efforts to develop specific inhibitors of the RPN2-RPN13 interaction, and suggest the existence of a previously unknown stress-response pathway.
- From the Departments of Biochemistry and.
Organizational Affiliation: