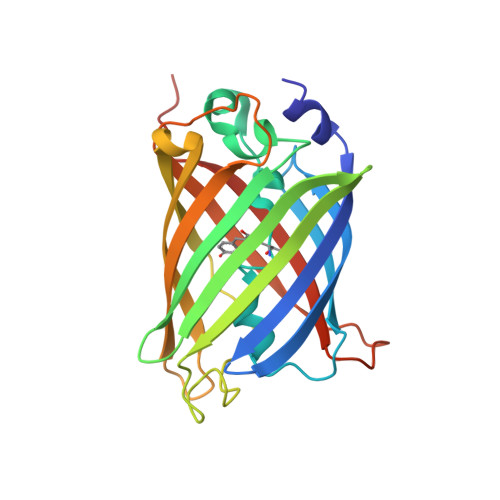
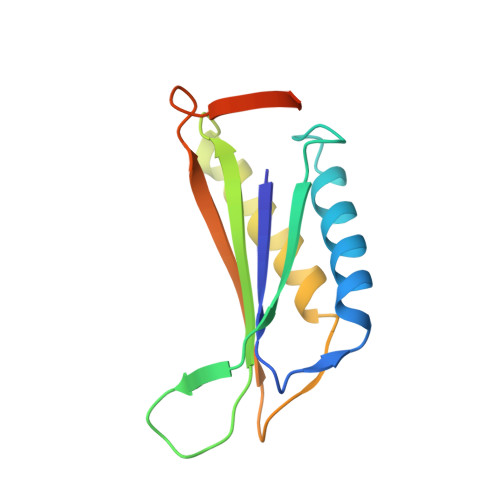
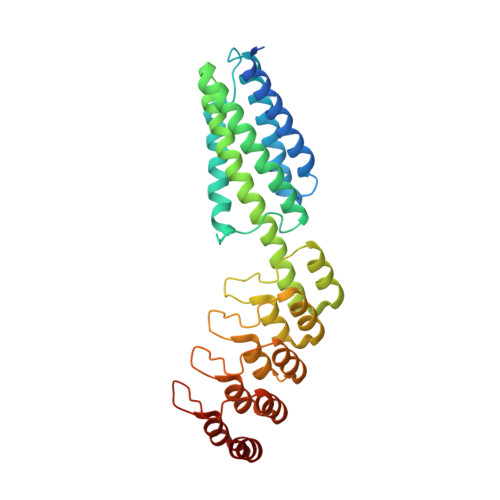
A 3.8 angstrom resolution cryo-EM structure of a small protein bound to an imaging scaffold.
Liu, Y., Huynh, D.T., Yeates, T.O.(2019) Nat Commun 10: 1864-1864
- PubMed: 31015551
- DOI: https://doi.org/10.1038/s41467-019-09836-0
- Primary Citation of Related Structures:
6NHT, 6NHV - PubMed Abstract:
Proteins smaller than about 50 kDa are currently too small to be imaged at high resolution by cryo-electron microscopy (cryo-EM), leaving most protein molecules in the cell beyond the reach of this powerful structural technique. Here we use a designed protein scaffold to bind and symmetrically display 12 copies of a small 26 kDa protein, green fluorescent protein (GFP). We show that the bound cargo protein is held rigidly enough to visualize it at a resolution of 3.8 Å by cryo-EM, where specific structural features of the protein are visible. The designed scaffold is modular and can be modified through modest changes in its amino acid sequence to bind and display diverse proteins for imaging, thus providing a general method to break through the lower size limitation in cryo-EM.
- UCLA Department of Chemistry and Biochemistry, Los Angeles, CA, 90095, USA.
Organizational Affiliation: