Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
Clinger, J.A., Elshahawi, S.I., Zhang, Y., Hall, R.P., Liu, Y., Miller, M.D., Thorson, J.S., Phillips Jr., G.N.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TerB Oxidoreductase | 334 | Streptomyces | Mutation(s): 0 | 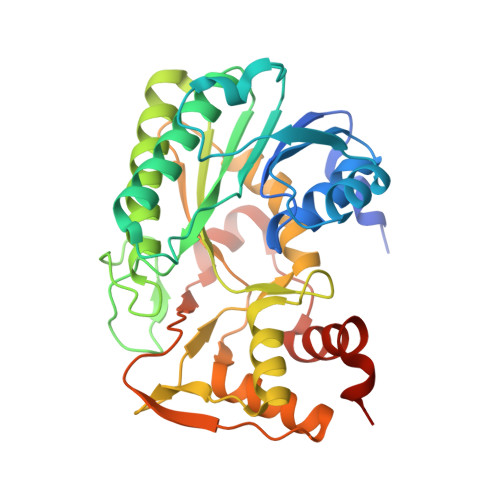 | |
UniProt | |||||
Find proteins for A0A3B6UEU0 (Streptomyces) Explore A0A3B6UEU0 Go to UniProtKB: A0A3B6UEU0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A3B6UEU0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NDP Query on NDP | C [auth A], G [auth B] | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE C21 H30 N7 O17 P3 ACFIXJIJDZMPPO-NNYOXOHSSA-N |  | ||
| KJG Query on KJG | D [auth A], H [auth B] | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione C18 H12 O4 HZKFHDXTSAYOSN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | E [auth A] F [auth A] I [auth B] J [auth B] K [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| IPA Query on IPA | N [auth B] | ISOPROPYL ALCOHOL C3 H8 O KFZMGEQAYNKOFK-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.498 | α = 90 |
| b = 80.991 | β = 90 |
| c = 139.955 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| SHELX | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM115261 |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | CA217255 |