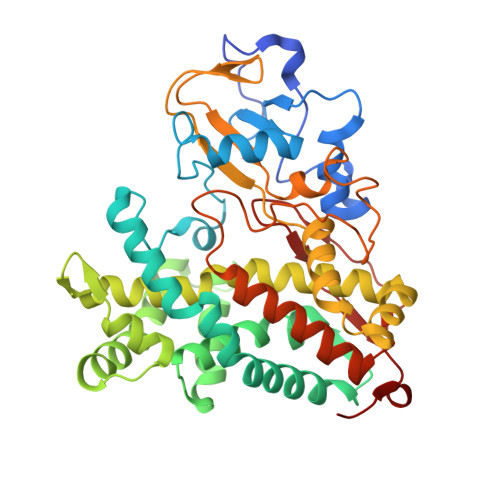
Ligand and Redox Partner Binding Generates a New Conformational State in Cytochrome P450cam (CYP101A1).
Follmer, A.H., Tripathi, S., Poulos, T.L.(2019) J Am Chem Soc 141: 2678-2683
- PubMed: 30672701
- DOI: https://doi.org/10.1021/jacs.8b13079
- Primary Citation of Related Structures:
6NBL - PubMed Abstract:
It has become increasingly clear that cytochromes P450 can cycle back and forth between two extreme conformational states termed the closed and open states. In the well-studied cytochrome P450cam, the binding of its redox partner, putidaredoxin (Pdx), shifts P450cam toward the open state. Shifting to the open state is thought to be important in the formation of a proton relay network essential for O-O bond cleavage and formation of the active Fe(IV)═O intermediate. Another important intermediate is the oxy-P450cam complex when bound to Pdx. Trapping this intermediate in crystallo is challenging owing to its instability, but the CN - complex is both stable and an excellent mimic of the O 2 complex. Here we present the P450cam-Pdx structure complexed with CN - . CN - results in large conformational changes including cis/trans isomerization of proline residues. Changes include large rearrangements of active-site residues and the formation of new active-site access channel that we have termed channel 2. The formation of channel 2 has also been observed in our previous molecular dynamics simulations wherein substrate binding to an allosteric site remote from the active site opens up channel 2. This new structure supports an extensive amount of previous work showing that distant regions of the structure are dynamically coupled and underscores the potentially important role that large conformational changes and dynamics play in P450 catalysis.
- Departments of Molecular Biology and Biochemistry, Pharmaceutical Sciences, and Chemistry , University of California , Irvine , California 92697-3900 , United States.
Organizational Affiliation: