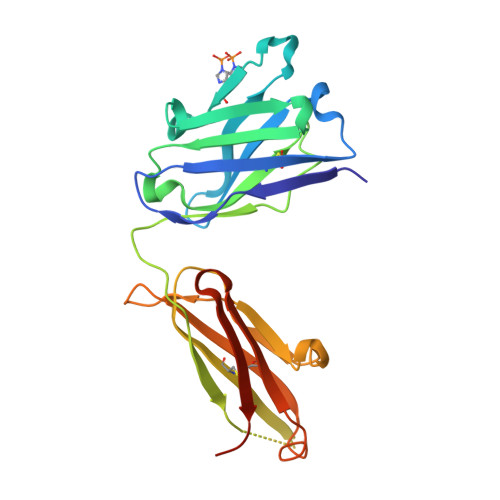
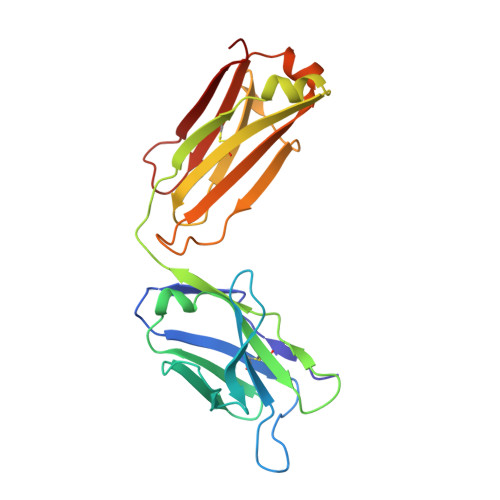
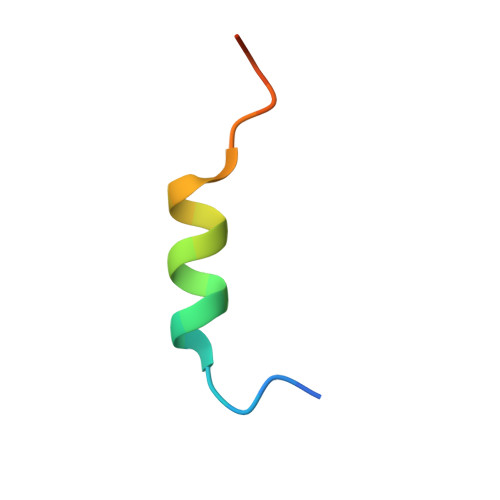
Identification of a Helical Segment within the Intrinsically Disordered Region of the PCSK9 Prodomain.
Ultsch, M., Li, W., Eigenbrot, C., Di Lello, P., Lipari, M.T., Gerhardy, S., AhYoung, A.P., Quinn, J., Franke, Y., Chen, Y., Kong Beltran, M., Peterson, A., Kirchhofer, D.(2019) J Mol Biology 431: 885-903
- PubMed: 30653992
- DOI: https://doi.org/10.1016/j.jmb.2018.11.025
- Primary Citation of Related Structures:
6E4Y, 6E4Z, 6MV5 - PubMed Abstract:
Proprotein convertase subtilisin/kexin 9 (PCSK9) is a key regulator of lipid metabolism by degrading liver LDL receptors. Structural studies have provided molecular details of PCSK9 function. However, the N-terminal acidic stretch of the PCSK9 prodomain (Q31-T60) has eluded structural investigation, since it is in a disordered state. The interest in this region is intensified by the presence of human missense mutations associated with low and high LDL-c levels (E32K, D35Y, and R46L, respectively), as well as two posttranslationally modified sites, sulfated Y38 and phosphorylated S47. Herein we show that a segment within this region undergoes disorder-to-order transition. Experiments with acidic stretch-derived peptides demonstrated that the folding is centered at the segment Y38-L45, which adopts an α-helix as determined by NMR analysis of free peptides and by X-ray crystallography of peptides in complex with antibody 6E2 (Ab6E2). In the Fab6E2-peptide complexes, the structured region features a central 2 1/4-turn α-helix and encompasses up to 2/3 of the length of the acidic stretch, including the missense mutations and posttranslationally modified sites. Experiments with helix-breaking proline substitutions in peptides and in PCSK9 protein indicated that Ab6E2 specifically recognizes the helical conformation of the acidic stretch. Therefore, the observed quantitative binding of Ab6E2 to native PCSK9 from various cell lines suggests that the disorder-to-order transition is a true feature of PCSK9 and not limited to peptides. Because the helix provides a constrained spatial orientation of the missense mutations and the posttranslationally modified residues, it is probable that their biological functions take place in the context of an ordered conformational state.
- Department of Structural Biology, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: