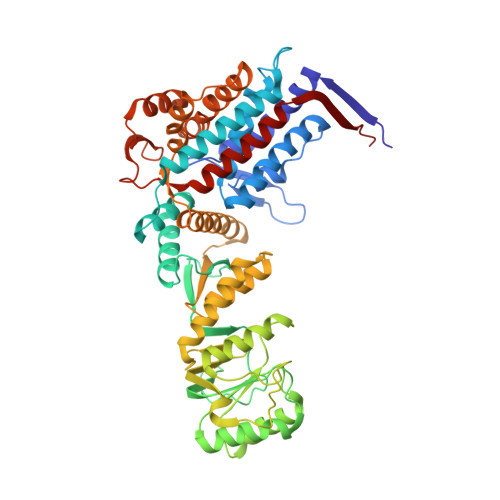
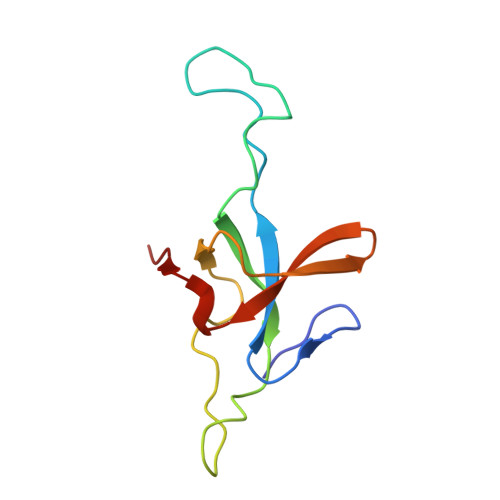
Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Gomez-Llorente, Y., Jebara, F., Patra, M., Malik, R., Nisemblat, S., Chomsky-Hecht, O., Parnas, A., Azem, A., Hirsch, J.A., Ubarretxena-Belandia, I.(2020) Nat Commun 11: 1916-1916
- PubMed: 32317635
- DOI: https://doi.org/10.1038/s41467-020-15698-8
- Primary Citation of Related Structures:
6MRC, 6MRD - PubMed Abstract:
mHsp60-mHsp10 assists the folding of mitochondrial matrix proteins without the negative ATP binding inter-ring cooperativity of GroEL-GroES. Here we report the crystal structure of an ATP (ADP:BeF 3 -bound) ground-state mimic double-ring mHsp60 14 -(mHsp10 7 ) 2 football complex, and the cryo-EM structures of the ADP-bound successor mHsp60 14 -(mHsp10 7 ) 2 complex, and a single-ring mHsp60 7 -mHsp10 7 half-football. The structures explain the nucleotide dependence of mHsp60 ring formation, and reveal an inter-ring nucleotide symmetry consistent with the absence of negative cooperativity. In the ground-state a two-fold symmetric H-bond and a salt bridge stitch the double-rings together, whereas only the H-bond remains as the equatorial gap increases in an ADP football poised to split into half-footballs. Refolding assays demonstrate obligate single- and double-ring mHsp60 variants are active, and complementation analysis in bacteria shows the single-ring variant is as efficient as wild-type mHsp60. Our work provides a structural basis for active single- and double-ring complexes coexisting in the mHsp60-mHsp10 chaperonin reaction cycle.
- Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, 10029, USA.
Organizational Affiliation: