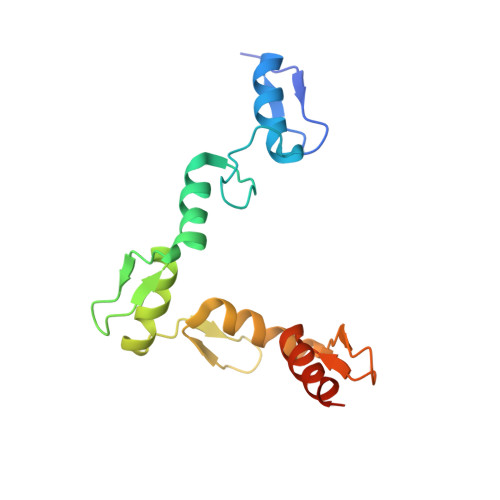
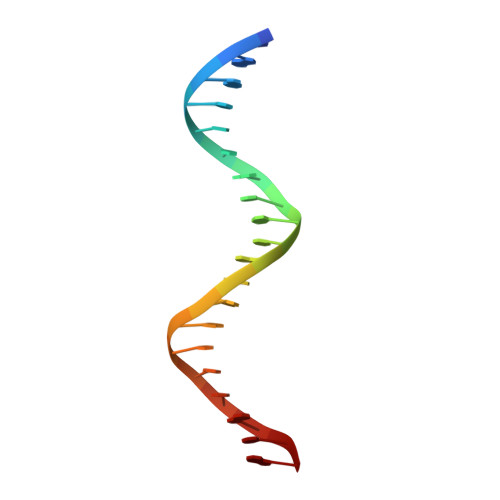
Structural basis of specific DNA binding by the transcription factor ZBTB24.
Ren, R., Hardikar, S., Horton, J.R., Lu, Y., Zeng, Y., Singh, A.K., Lin, K., Coletta, L.D., Shen, J., Lin Kong, C.S., Hashimoto, H., Zhang, X., Chen, T., Cheng, X.(2019) Nucleic Acids Res 47: 8388-8398
- PubMed: 31226215
- DOI: https://doi.org/10.1093/nar/gkz557
- Primary Citation of Related Structures:
6ML2, 6ML3, 6ML4, 6ML5, 6ML6, 6ML7 - PubMed Abstract:
ZBTB24, encoding a protein of the ZBTB family of transcriptional regulators, is one of four known genes-the other three being DNMT3B, CDCA7 and HELLS-that are mutated in immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome, a genetic disorder characterized by DNA hypomethylation and antibody deficiency. The molecular mechanisms by which ZBTB24 regulates gene expression and the biological functions of ZBTB24 are poorly understood. Here, we identified a 12-bp consensus sequence [CT(G/T)CCAGGACCT] occupied by ZBTB24 in the mouse genome. The sequence is present at multiple loci, including the Cdca7 promoter region, and ZBTB24 binding is mostly associated with gene activation. Crystallography and DNA-binding data revealed that the last four of the eight zinc fingers (ZFs) (i.e. ZF5-8) in ZBTB24 confer specificity of DNA binding. Two ICF missense mutations have been identified in the ZBTB24 ZF domain, which alter zinc-binding cysteine residues. We demonstrated that the corresponding C382Y and C407G mutations in mouse ZBTB24 abolish specific DNA binding and fail to induce Cdca7 expression. Our analyses indicate and suggest a structural basis for the sequence specific recognition by a transcription factor centrally important for the pathogenesis of ICF syndrome.
- Department of Epigenetics and Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Organizational Affiliation: