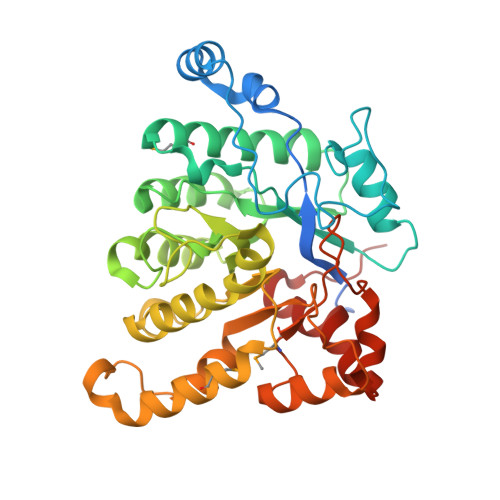
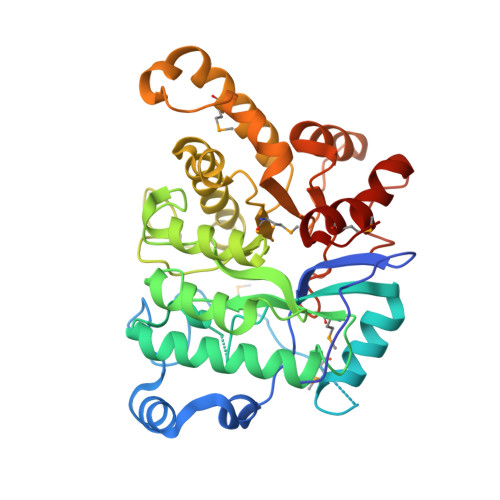
A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Hibi, M., Fukuda, D., Kenchu, C., Nojiri, M., Hara, R., Takeuchi, M., Aburaya, S., Aoki, W., Mizutani, K., Yasohara, Y., Ueda, M., Mikami, B., Takahashi, S., Ogawa, J.(2021) Commun Biol 4: 16-16
- PubMed: 33398074
- DOI: https://doi.org/10.1038/s42003-020-01555-3
- Primary Citation of Related Structures:
6M1W, 6M2I - PubMed Abstract:
The high-valent iron-oxo species formed in the non-heme diiron enzymes have high oxidative reactivity and catalyze difficult chemical reactions. Although the hydroxylation of inert methyl groups is an industrially promising reaction, utilizing non-heme diiron enzymes as such a biocatalyst has been difficult. Here we show a three-component monooxygenase system for the selective terminal hydroxylation of α-aminoisobutyric acid (Aib) into α-methyl-D-serine. It consists of the hydroxylase component, AibH1H2, and the electron transfer component. Aib hydroxylation is the initial step of Aib catabolism in Rhodococcus wratislaviensis C31-06, which has been fully elucidated through a proteome analysis. The crystal structure analysis revealed that AibH1H2 forms a heterotetramer of two amidohydrolase superfamily proteins, of which AibHm2 is a non-heme diiron protein and functions as a catalytic subunit. The Aib monooxygenase was demonstrated to be a promising biocatalyst that is suitable for bioprocesses in which the inert C-H bond in methyl groups need to be activated.
- Biotechnology Research Center and Department of Biotechnology, Toyama Prefectural University, 5180 Kurokawa, Imizu, Toyama, 939-0398, Japan. hibi@pu-toyama.ac.jp.
Organizational Affiliation: