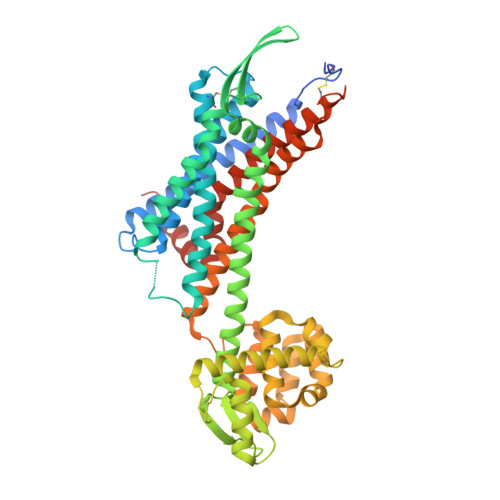
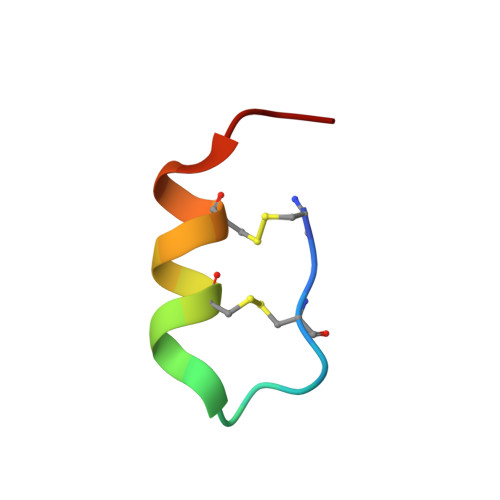
Crystal structure of human endothelin ETBreceptor in complex with sarafotoxin S6b.
Izume, T., Miyauchi, H., Shihoya, W., Nureki, O.(2020) Biochem Biophys Res Commun 528: 383-388
- PubMed: 32001000
- DOI: https://doi.org/10.1016/j.bbrc.2019.12.091
- Primary Citation of Related Structures:
6LRY - PubMed Abstract:
Sarafotoxins (SRTXs) are endothelin-like peptides extracted from snake venom. SRTXs stimulate the endothelin ET A and ET B receptors and enhance vasoconstriction, followed by left ventricular dysfunction and bronchoconstriction. SRTXs include four major isopeptides, S6a-d, with different subtype selectivities. Here, we report the crystal structure of the human ET B receptor in complex with the non-selective sarafotoxin S6b at 3.0 Å resolution. This structure reveals the similarities and differences between the binding modes of the endothelins and S6b. Moreover, molecular dynamics simulations based on the S6b-bound receptor provides structural insight into the subtype selectivity of the sarafotoxins. Our study clarifies the recognition mechanism of the endothelin-like peptide families.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo, Tokyo, 113-0033, Japan.
Organizational Affiliation: