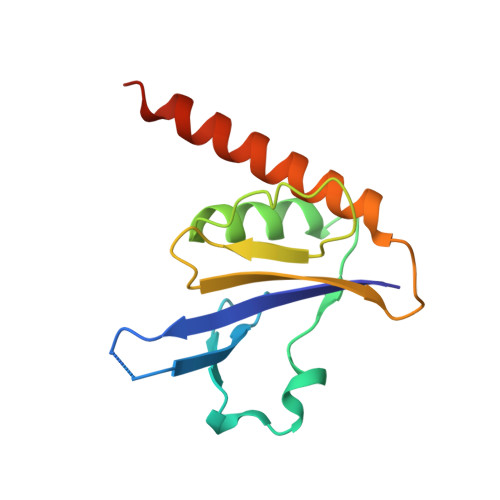
Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Wang, Y., Jin, H., Yang, F., Jiang, Y.L., Zhao, Y.Y., Chen, Z.P., Li, W.F., Chen, Y., Zhou, C.Z., Li, Q.(2020) Proteins 88: 1226-1232
- PubMed: 32337767
- DOI: https://doi.org/10.1002/prot.25896
- Primary Citation of Related Structures:
6L2W - PubMed Abstract:
Cyanophages, widespread in aquatic systems, are a class of viruses that specifically infect cyanobacteria. Though they play important roles in modulating the homeostasis of cyanobacterial populations, little is known about the freshwater cyanophages, especially those hypothetical proteins of unknown function. Mic1 is a freshwater siphocyanophage isolated from the Lake Chaohu. It encodes three hypothetical proteins Gp65, Gp66, and Gp72, which share an identity of 61.6% to 83%. However, we find these three homologous proteins differ from each other in oligomeric state. Moreover, we solve the crystal structure of Gp72 at 2.3 Å, which represents a novel fold in the α + β class. Structural analyses combined with redox assays enable us to propose a model of disulfide bond mediated oligomerization for Gp72. Altogether, these findings provide structural and biochemical basis for further investigations on the freshwater cyanophage Mic1.
- Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China.
Organizational Affiliation: