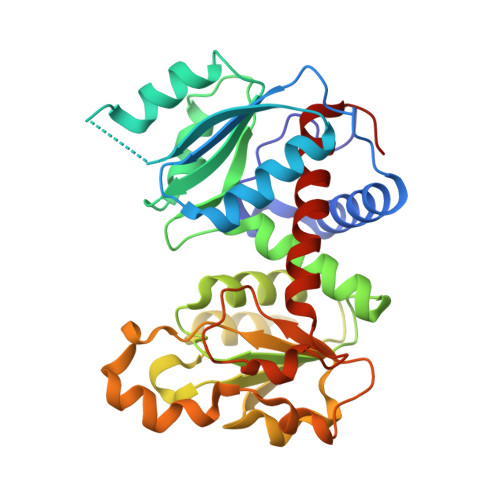
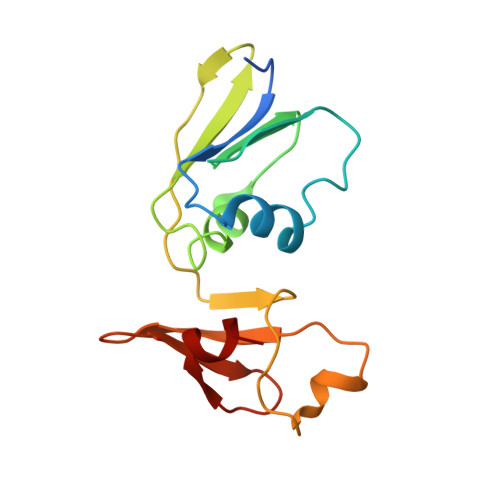
Conformational Plasticity of the Active Site Entrance inE. coliAspartate Transcarbamoylase and Its Implication in Feedback Regulation.
Lei, Z., Wang, N., Tan, H., Zheng, J., Jia, Z.(2020) Int J Mol Sci 21
- PubMed: 31947715
- DOI: https://doi.org/10.3390/ijms21010320
- Primary Citation of Related Structures:
6KJB - PubMed Abstract:
Aspartate transcarbamoylase (ATCase) has been studied for decades and Escherichia coli ATCase is referred as a "textbook example" for both feedback regulation and cooperativity. However, several critical questions about the catalytic and regulatory mechanisms of E. coli ATCase remain unanswered, especially about its remote feedback regulation. Herein, we determined a structure of E. coli ATCase in which a key residue located (Arg167) at the entrance of the active site adopted an uncommon open conformation, representing the first wild-type apo-form E. coli ATCase holoenzyme that features this state. Based on the structure and our results of enzymatic characterization, as well as molecular dynamic simulations, we provide new insights into the feedback regulation of E. coli ATCase. We speculate that the binding of pyrimidines or purines would affect the hydrogen bond network at the interface of the catalytic and regulatory subunit, which would further influence the stability of the open conformation of Arg167 and the enzymatic activity of ATCase. Our results not only revealed the importance of the previously unappreciated open conformation of Arg167 in the active site, but also helped to provide rationalization for the mechanism of the remote feedback regulation of ATCase.
- College of Chemistry, Beijing Normal University, Beijing 100875, China.
Organizational Affiliation: