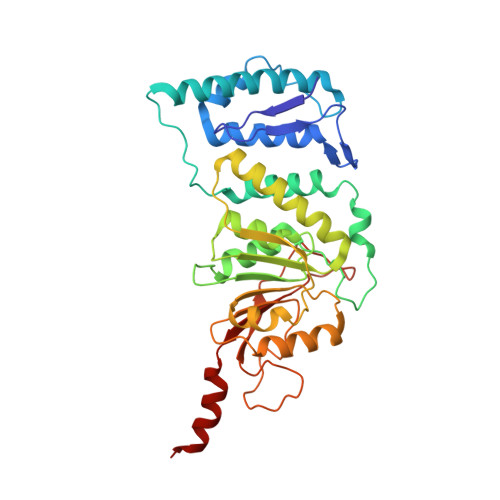
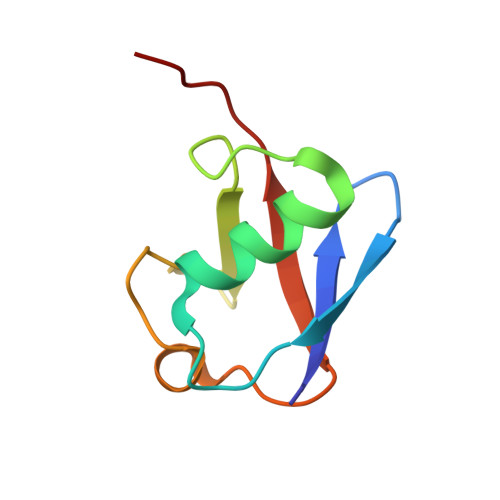
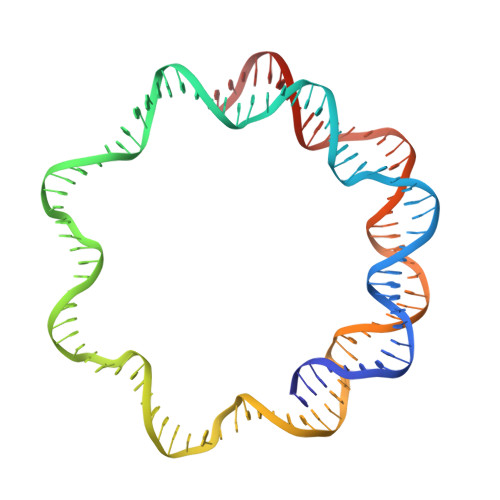
Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Jang, S., Kang, C., Yang, H.S., Jung, T., Hebert, H., Chung, K.Y., Kim, S.J., Hohng, S., Song, J.J.(2019) Genes Dev 33: 620-625
- PubMed: 30923167
- DOI: https://doi.org/10.1101/gad.323790.118
- Primary Citation of Related Structures:
6JM9, 6JMA - PubMed Abstract:
DOT1L is a histone H3 Lys79 methyltransferase whose activity is stimulated by histone H2B Lys120 ubiquitination, suggesting cross-talk between histone H3 methylation and H2B ubiquitination. Here, we present cryo-EM structures of DOT1L complexes with unmodified or H2B ubiquitinated nucleosomes, showing that DOT1L recognizes H2B ubiquitin and the H2A/H2B acidic patch through a C-terminal hydrophobic helix and an arginine anchor in DOT1L, respectively. Furthermore, the structures combined with single-molecule FRET experiments show that H2B ubiquitination enhances a noncatalytic function of the DOT1L-destabilizing nucleosome. These results establish the molecular basis of the cross-talk between H2B ubiquitination and H3 Lys79 methylation as well as nucleosome destabilization by DOT1L.
- Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon 34141, Korea.
Organizational Affiliation: