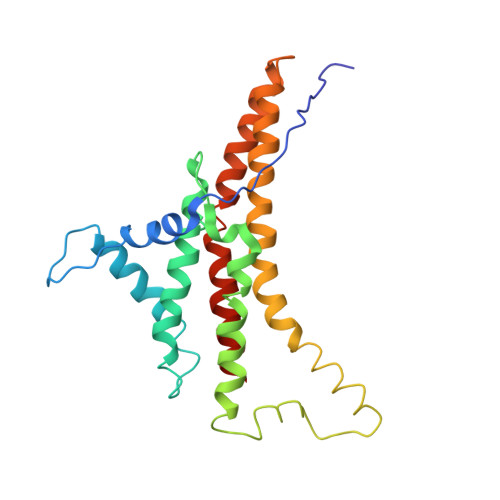
Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Gu, J., Zhang, L., Zong, S., Guo, R., Liu, T., Yi, J., Wang, P., Zhuo, W., Yang, M.(2019) Science 364: 1068-1075
- PubMed: 31197009
- DOI: https://doi.org/10.1126/science.aaw4852
- Primary Citation of Related Structures:
6J54, 6J5A, 6J5I, 6J5J, 6J5K - PubMed Abstract:
The mitochondrial adenosine triphosphate (ATP) synthase produces most of the ATP required by mammalian cells. We isolated porcine tetrameric ATP synthase and solved its structure at 6.2-angstrom resolution using a single-particle cryo-electron microscopy method. Two classical V-shaped ATP synthase dimers lie antiparallel to each other to form an H-shaped ATP synthase tetramer, as viewed from the matrix. ATP synthase inhibitory factor subunit 1 (IF1) is a well-known in vivo inhibitor of mammalian ATP synthase at low pH. Two IF1 dimers link two ATP synthase dimers, which is consistent with the ATP synthase tetramer adopting an inhibited state. Within the tetramer, we refined structures of intact ATP synthase in two different rotational conformations at 3.34- and 3.45-Å resolution.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: