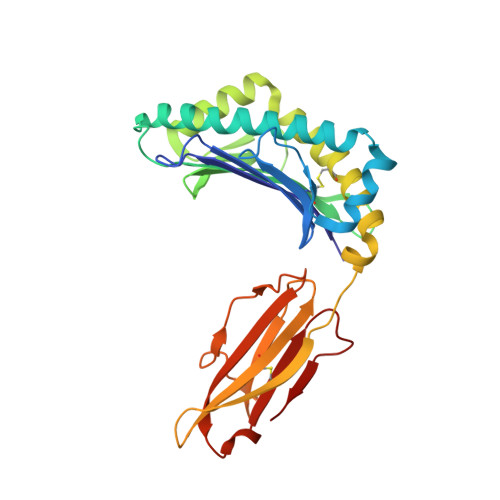
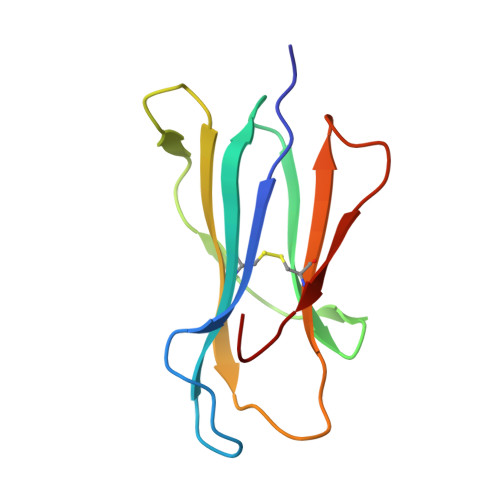
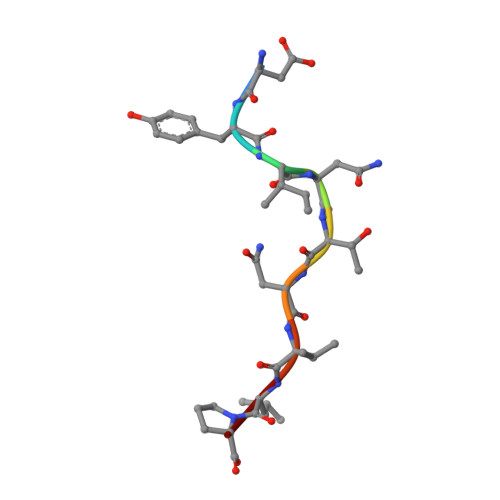
Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Lu, D., Liu, K., Zhang, D., Yue, C., Lu, Q., Cheng, H., Wang, L., Chai, Y., Qi, J., Wang, L.F., Gao, G.F., Liu, W.J.(2019) PLoS Biol 17: e3000436-e3000436
- PubMed: 31498797
- DOI: https://doi.org/10.1371/journal.pbio.3000436
- Primary Citation of Related Structures:
6J2D, 6J2E, 6J2F, 6J2G, 6J2H, 6J2I, 6J2J, 6K7T, 6K7U - PubMed Abstract:
Bats harbor many zoonotic viruses, including highly pathogenic viruses of humans and other mammals, but they are typically asymptomatic in bats. To further understand the antiviral immunity of bats, we screened and identified a series of bat major histocompatibility complex (MHC) I Ptal-N*01:01-binding peptides derived from four different bat-borne viruses, i.e., Hendra virus (HeV), Ebola virus (EBOV), Middle East respiratory syndrome coronavirus (MERS-CoV), and H17N10 influenza-like virus. The structures of Ptal-N*01:01 display unusual peptide presentation features in that the bat-specific 3-amino acid (aa) insertion enables the tight "surface anchoring" of the P1-Asp in pocket A of bat MHC I. As the classical primary anchoring positions, the B and F pockets of Ptal-N*01:01 also show unconventional conformations, which contribute to unusual peptide motifs and distinct peptide presentation. Notably, the features of bat MHC I may be shared by MHC I from various marsupials. Our study sheds light on bat adaptive immunity and may benefit future vaccine development against bat-borne viruses of high impact on humans.
- NHC Key Laboratory of Medical Virology and Viral Diseases, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China.
Organizational Affiliation: