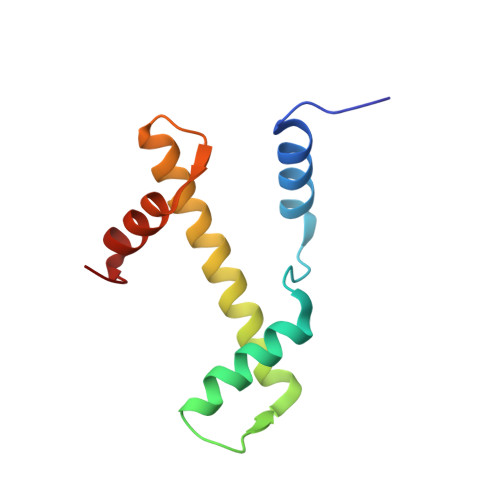
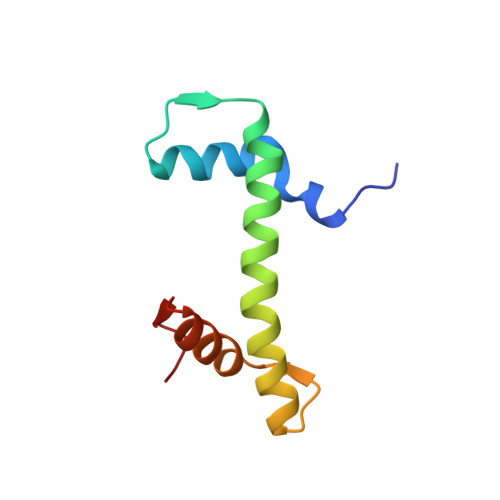
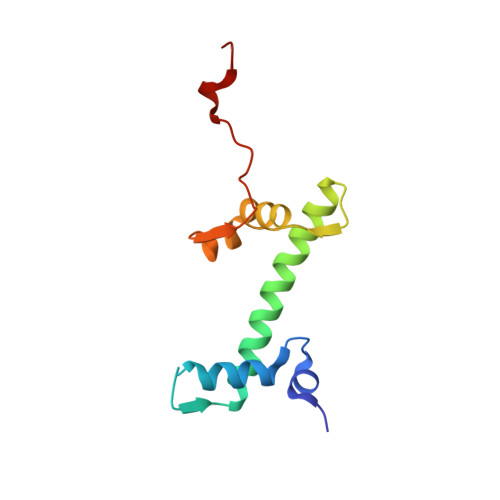
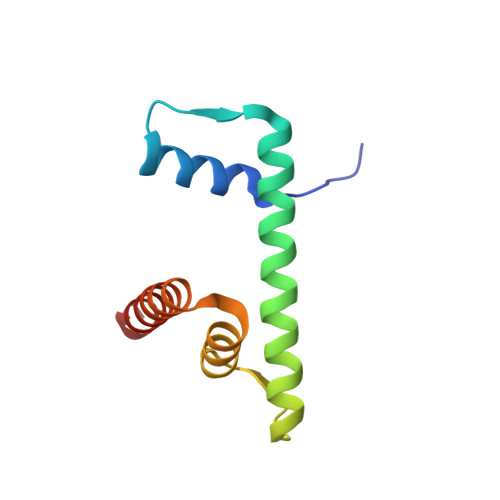
Crosslinking Allosteric Sites on the Nucleosome.
Batchelor, L.K., De Falco, L., von Erlach, T., Sharma, D., Adhireksan, Z., Roethlisberger, U., Davey, C.A., Dyson, P.J.(2019) Angew Chem Int Ed Engl 58: 15660-15664
- PubMed: 31478581
- DOI: https://doi.org/10.1002/anie.201906423
- Primary Citation of Related Structures:
6IQ4 - PubMed Abstract:
Targeting defined histone protein sites in chromatin is an emerging therapeutic approach that can potentially be enhanced by allosteric effects within the nucleosome. Here we characterized a novel hetero-bimetallic compound with a design based on a nucleosomal allostery effect observed earlier for two unrelated drugs-the Ru II antimetastasis/antitumor RAPTA-T and the Au I anti-arthritic auranofin. The Ru II moiety binds specifically to two H2A glutamate residues on the nucleosome acidic patch, allosterically triggering a cascade of structural changes that promote binding of the Au I moiety to selective histidine residues on H3, resulting in cross-linking sites that are over 35 Å distant. By tethering the H2A-H2B dimers to the H3-H4 tetramer, the hetero-bimetallic compound significantly increases stability of the nucleosome, illustrating its utility as a site-selective cross-linking agent.
- Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland.
Organizational Affiliation: