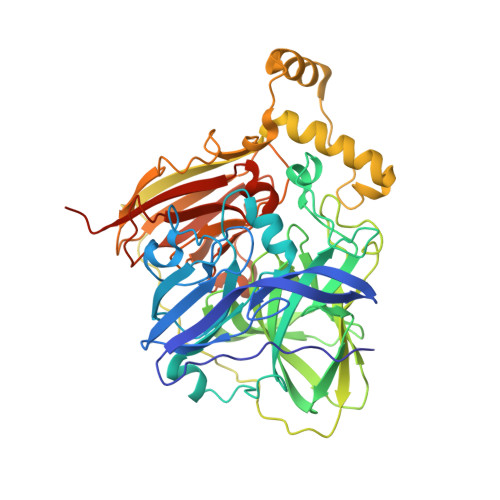
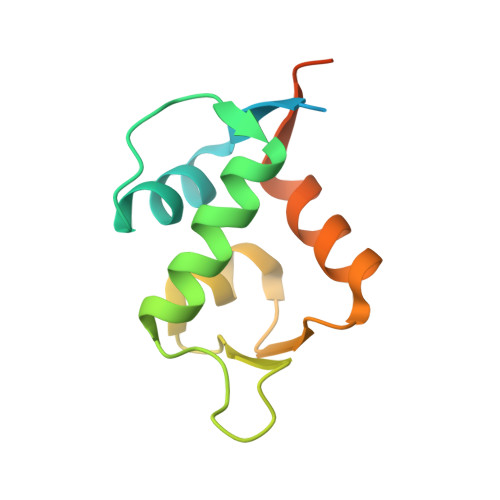
Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
Sana, B., Chee, S.M.Q., Wongsantichon, J., Raghavan, S., Robinson, R.C., Ghadessy, F.J.(2019) J Biological Chem 294: 7002-7012
- PubMed: 30770473
- DOI: https://doi.org/10.1074/jbc.RA118.007141
- Primary Citation of Related Structures:
6IM7, 6IM8, 6IM9 - PubMed Abstract:
Protein-protein interactions (PPIs) are ubiquitous in almost all biological processes and are often corrupted in diseased states. A detailed understanding of PPIs is therefore key to understanding cellular physiology and can yield attractive therapeutic targets. Here, we describe the development and structural characterization of novel Escherichia coli CueO multi-copper oxidase variants engineered to recapitulate protein-protein interactions with commensurate modulation of their enzymatic activities. The fully integrated single-protein sensors were developed through modular grafting of ligand-specific peptides into a highly compliant and flexible methionine-rich loop of CueO. Sensitive detection of diverse ligand classes exemplified by antibodies, an E3 ligase, MDM2 proto-oncogene (MDM2), and protease (SplB from Staphylococcus aureus ) was achieved in a simple mix and measure homogeneous format with visually observable colorimetric readouts. Therapeutic antagonism of MDM2 by small molecules and peptides in clinical development for treatment of cancer patients was assayed using the MDM2-binding CueO enzyme. Structural characterization of the free and MDM2-bound CueO variant provided functional insight into signal-transducing mechanisms of the engineered enzymes and highlighted the robustness of CueO as a stable and compliant scaffold for multiple applications.
- From the p53 Laboratory, Agency for Science, Technology, and Research (A*STAR), 8A Biomedical Grove, Singapore 138648, Singapore.
Organizational Affiliation: